 |  |
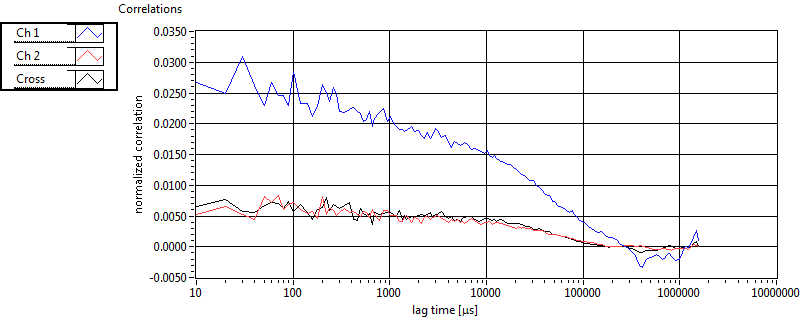
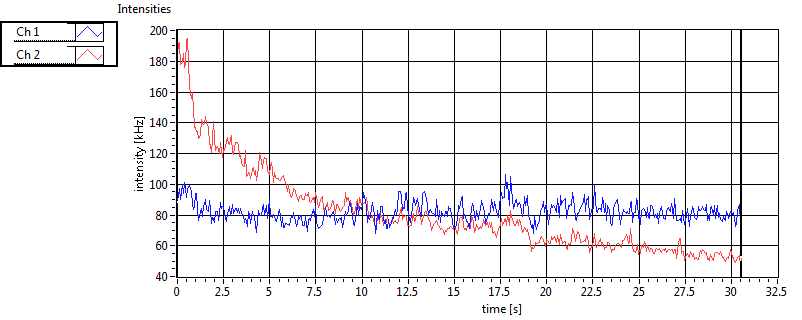
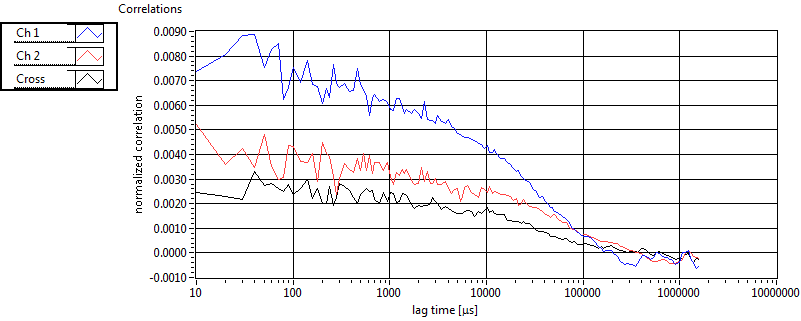
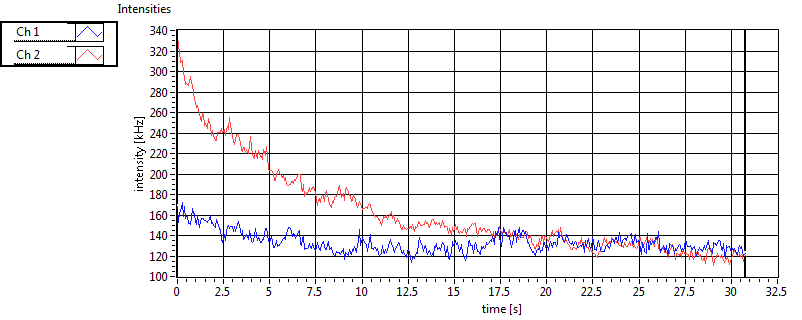
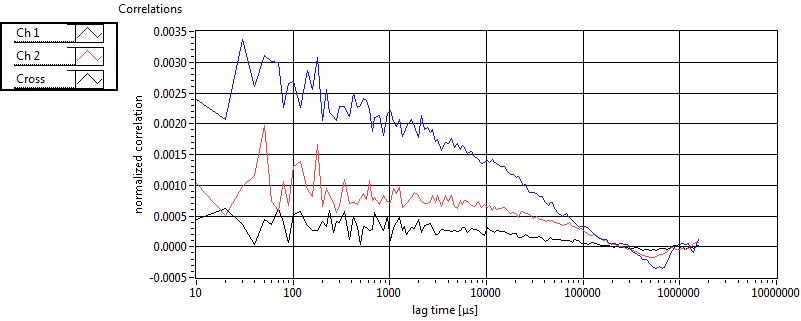
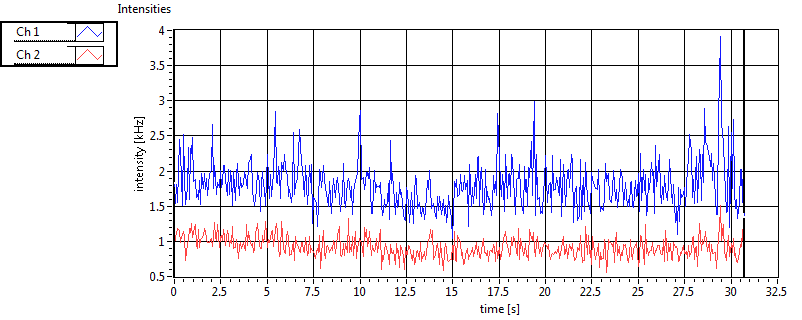
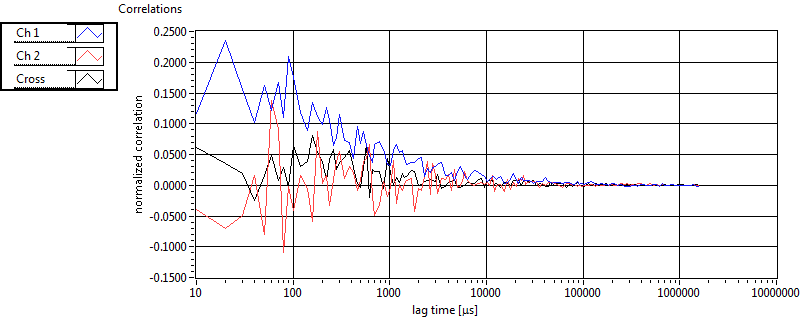
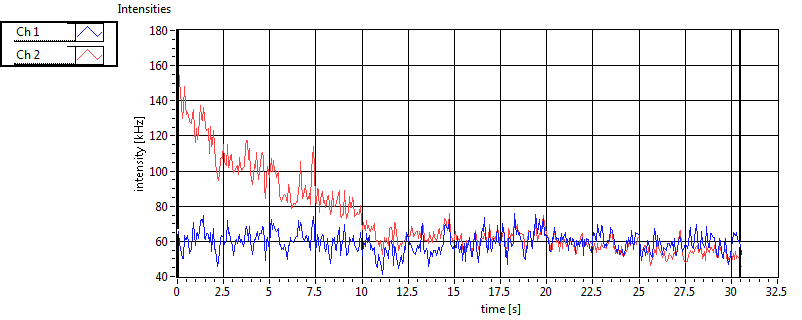
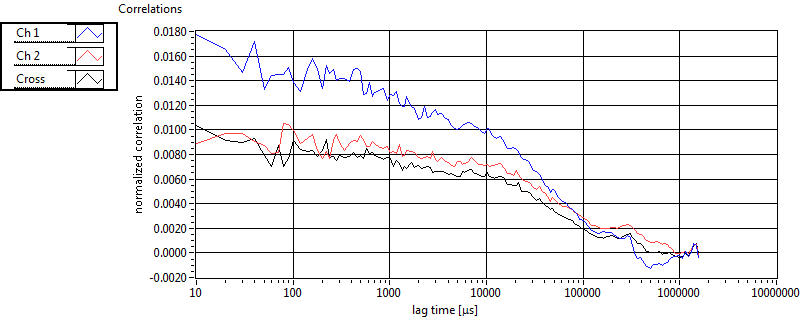
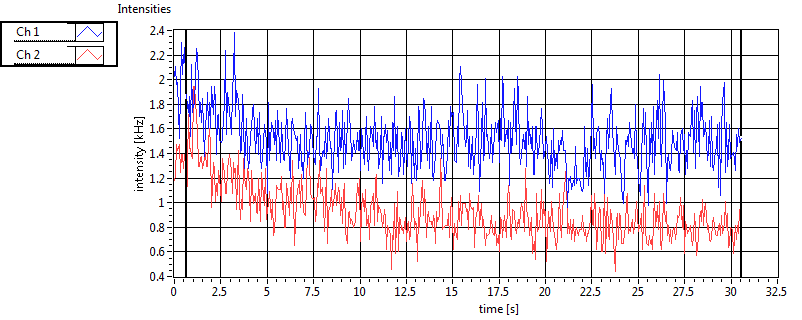
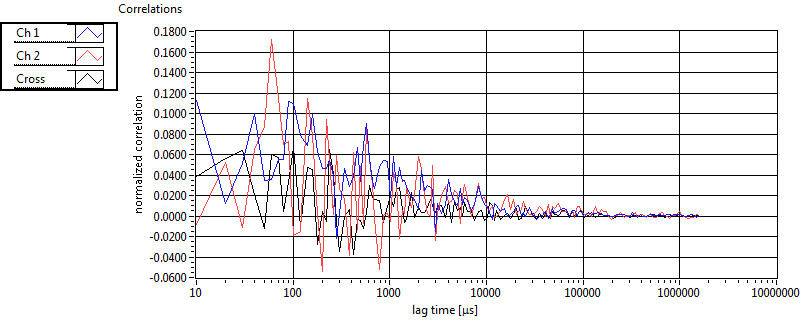
| 1 | 20130125\U03--V00--3155hi |
|---|---|
| 2 | 20130125\U03--V01--3155hi |
| 3 | 20130131\U02--V00--3155hi |
| 4 | 20130131\U02--V01--3155hi |
| 5 | 20130201\U02--V00--3155hi |
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 1.553E+2 | 9.835E+1 | 6.609E+2 | 3.163E+2 | 5.245E+1 | 5.560E+1 | 4.521E+1 | 8.043E+1 | 2.731E+2 | 1.220E+2 | 2.227E+1 | 8.018E+0 |
| N_corr | 1.749E+2 | 1.225E+2 | 1.074E+3 | 5.562E+2 | 7.497E+1 | 9.029E+1 | 4.029E+1 | 8.134E+1 | 3.863E+2 | 2.224E+2 | 2.406E+1 | 1.021E+1 |
| c [nM] | 9.661E+2 | 6.117E+2 | 3.065E+3 | 1.467E+3 | 2.696E+2 | 2.858E+2 | 2.812E+2 | 5.002E+2 | 1.267E+3 | 5.661E+2 | 1.145E+2 | 4.122E+1 |
| c_corr [nM] | 1.088E+3 | 7.616E+2 | 4.984E+3 | 2.580E+3 | 3.854E+2 | 4.642E+2 | 2.506E+2 | 5.059E+2 | 1.792E+3 | 1.032E+3 | 1.237E+2 | 5.247E+1 |
| ratioG | 2.293E-1 | 1.137E-1 | 2.888E-1 | 1.436E-1 | ||||||||
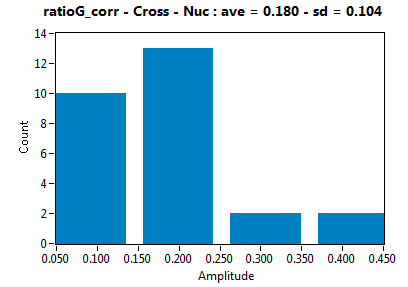
| ratioG_corr | 1.798E-1 | 1.035E-1 | 2.537E-1 | 1.256E-1 | ||||||||
| f_1 | 6.523E-1 | 2.171E-1 | 2.708E-1 | 1.691E-1 | 8.199E-1 | 1.922E-1 | 8.463E-1 | 1.943E-1 | 4.628E-1 | 2.003E-1 | 8.163E-1 | 2.172E-1 |
| tauD_1 [ms] | 6.143E+0 | 5.338E+0 | 1.860E+0 | 0.000E+0 | 9.744E+0 | 8.992E+0 | 3.187E+0 | 7.137E+0 | 1.860E+0 | 0.000E+0 | 1.496E+1 | 1.225E+1 |
| D_1 [mum^2/s] | 4.224E+0 | 4.097E+0 | 6.926E+0 | 0.000E+0 | 3.115E+0 | 3.490E+0 | 2.367E+1 | 2.286E+1 | 6.926E+0 | 0.000E+0 | 1.787E+0 | 1.443E+0 |
| alpha_1 | 9.457E-1 | 1.113E-1 | 9.200E-1 | 1.110E-16 | 9.102E-1 | 2.017E-1 | 1.044E+0 | 1.705E-1 | 9.200E-1 | 0.000E+0 | 1.103E+0 | 1.375E-1 |
| f_2 | 3.477E-1 | 2.171E-1 | 7.292E-1 | 1.691E-1 | 1.801E-1 | 1.922E-1 | 1.537E-1 | 1.943E-1 | 5.372E-1 | 2.003E-1 | 1.837E-1 | 2.172E-1 |
| tauD_2 [ms] | 6.972E+1 | 4.348E+1 | 5.580E+1 | 3.223E+1 | 1.343E+2 | 7.534E+1 | 2.950E+1 | 2.697E+1 | 6.427E+1 | 8.129E+0 | 1.452E+2 | 4.683E+1 |
| D_2 [mum^2/s] | 1.912E-1 | 8.210E-2 | 3.503E-1 | 3.222E-1 | 1.200E-1 | 6.684E-2 | 6.085E-1 | 3.572E-1 | 2.041E-1 | 2.879E-2 | 9.397E-2 | 3.598E-2 |
| alpha_2 | 1.506E+0 | 3.401E-1 | 1.396E+0 | 3.388E-1 | 1.466E+0 | 4.304E-1 | 1.219E+0 | 2.841E-1 | 1.390E+0 | 3.880E-1 | 1.395E+0 | 4.283E-1 |
| tauD_m [ms] | 2.515E+1 | 1.103E+1 | 3.978E+1 | 2.355E+1 | 2.364E+1 | 1.344E+1 | 8.204E+0 | 1.187E+1 | 3.608E+1 | 1.504E+1 | 3.143E+1 | 1.525E+1 |
| D_m [mum^2/s] | 2.213E+0 | 1.659E+0 | 2.152E+0 | 1.042E+0 | 2.258E+0 | 2.252E+0 | 2.216E+1 | 2.269E+1 | 3.312E+0 | 1.355E+0 | 1.190E+0 | 6.701E-1 |
| R [1/s] | 4.046E+0 | 7.906E+0 | 1.190E+0 | 1.683E+0 | ||||||||
| CPM [kHz] | 4.016E-1 | 1.804E-1 | 3.263E-1 | 2.815E-1 | 2.250E-1 | 1.386E-1 | 5.178E-1 | 3.635E-1 | ||||
| CPM_corr [kHz] | 4.183E-1 | 1.784E-1 | 3.644E-1 | 3.027E-1 | 8.811E+0 | 4.374E+1 | 5.756E-1 | 4.313E-1 | ||||
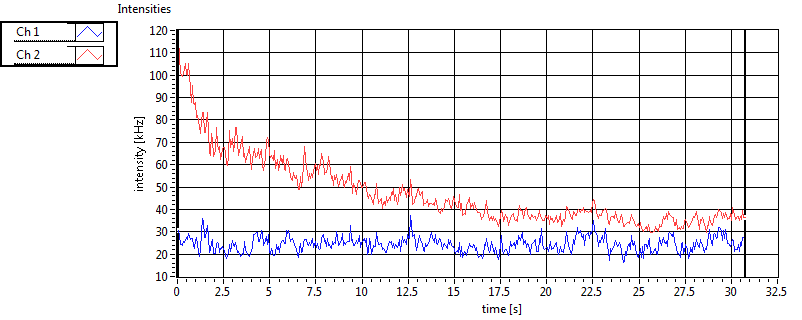
| Int [kHz] | 6.561E+1 | 6.331E+1 | 1.151E+2 | 3.221E+1 | 1.105E+1 | 1.737E+1 | 7.222E+1 | 1.493E+1 | ||||
| Int_corr [kHz] | 7.769E+1 | 8.348E+1 | 2.105E+2 | 7.229E+1 | 1.170E+1 | 1.827E+1 | 1.093E+2 | 3.879E+1 | ||||
| # samples | 59 | 59 | 46 | 46 | 27 | 27 | 39 | 39 | 4 | 4 | 3 | 3 |
| # cells | 30 |
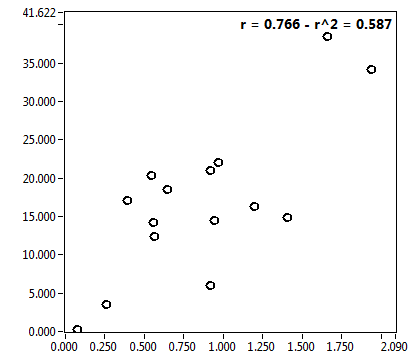
![c_corr [nM] - Ch1 - Nuc : ave = 1087.651 - sd = 761.556](3155hi_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 77.694 - sd = 83.483](3155hi_Int_corr_1n.png)
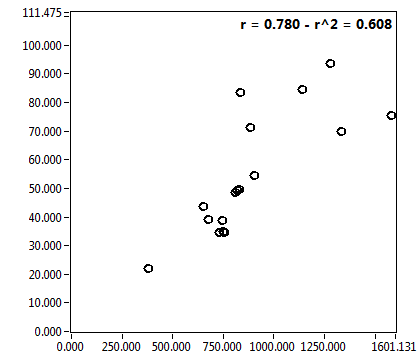
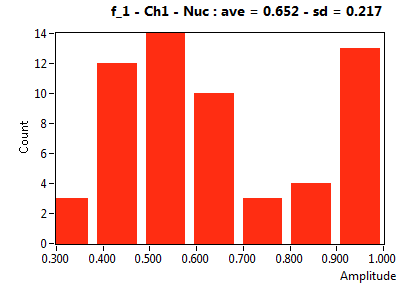
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 4.224 - sd = 4.097](3155hi_D_1_1n.png)
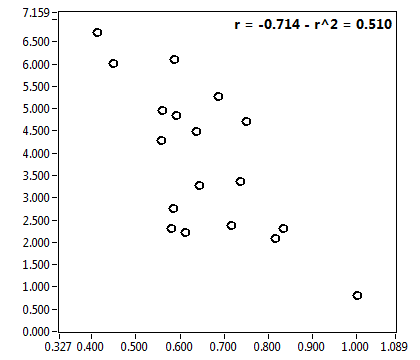
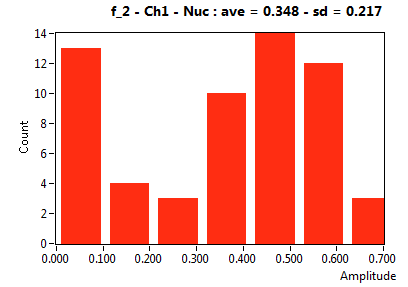
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.191 - sd = 0.082](3155hi_D_2_1n.png)
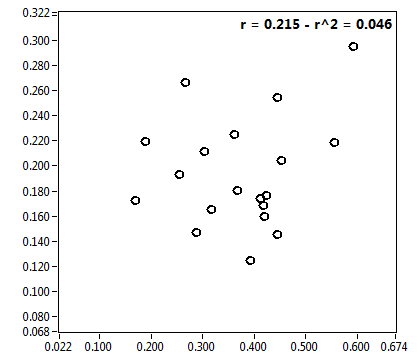
![CPM_corr [kHz] - Ch1 - Nuc : ave = 0.418 - sd = 0.178](3155hi_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 2.213 - sd = 1.659](3155hi_D_m_1n.png)
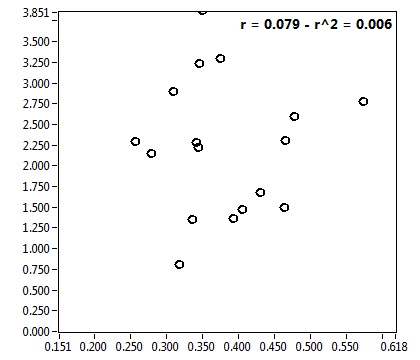
![c_corr [nM] - Ch1 - Nuc : ave = 1087.651 - sd = 761.556](3155hi_c_corr_1n.png)
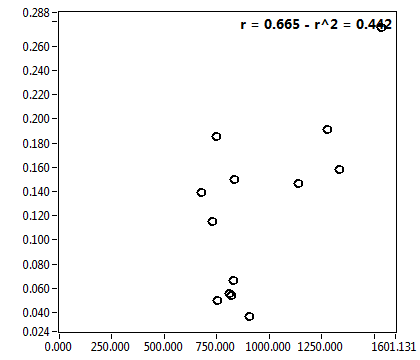
![c_corr [nM] - Ch1 - Cyto : ave = 250.554 - sd = 505.899](3155hi_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 11.703 - sd = 18.267](3155hi_Int_corr_1c.png)
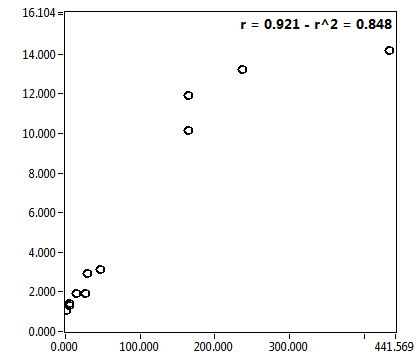
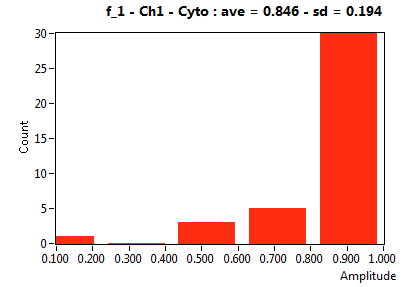
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 23.671 - sd = 22.863](3155hi_D_1_1c.png)
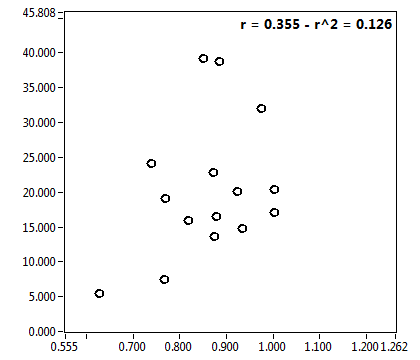
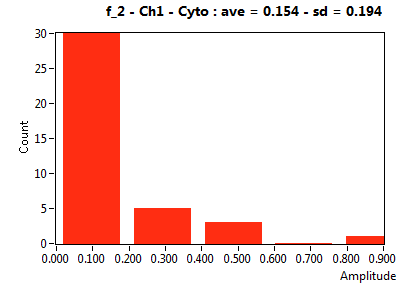
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 0.608 - sd = 0.357](3155hi_D_2_1c.png)
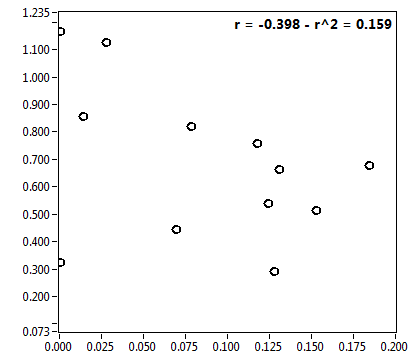
![CPM_corr [kHz] - Ch1 - Cyto : ave = 8.811 - sd = 43.737](3155hi_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 22.163 - sd = 22.688](3155hi_D_m_1c.png)