 |  |
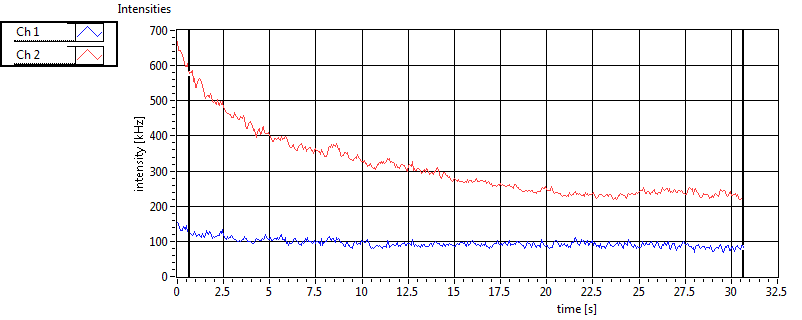
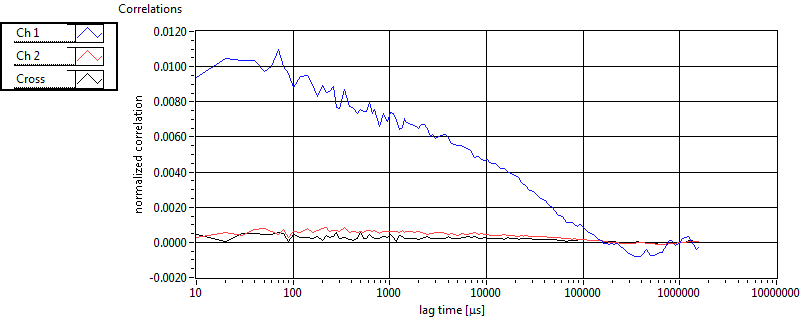
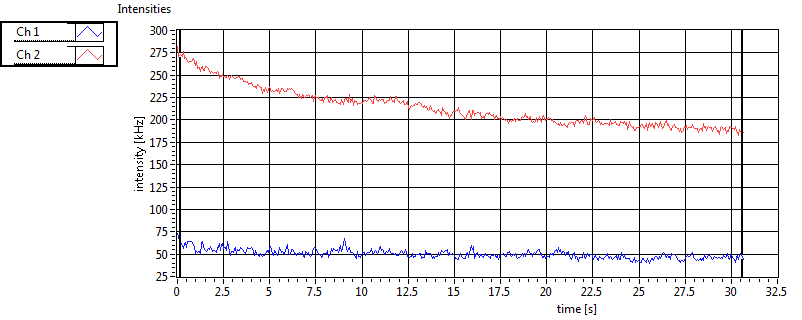
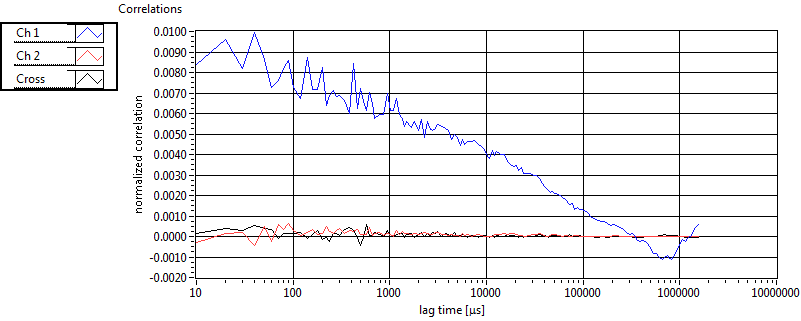
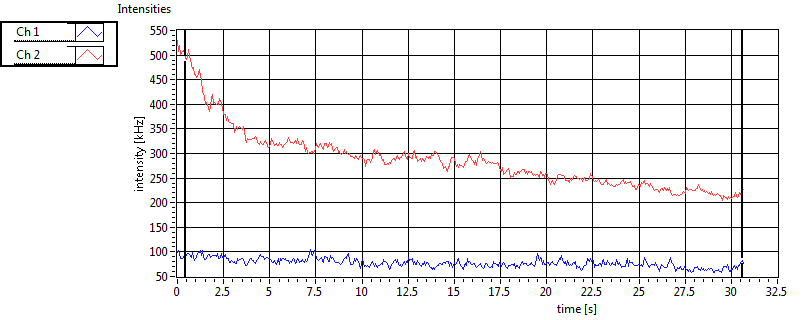
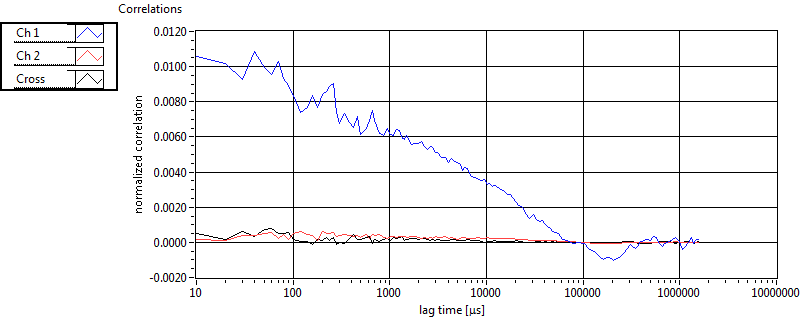
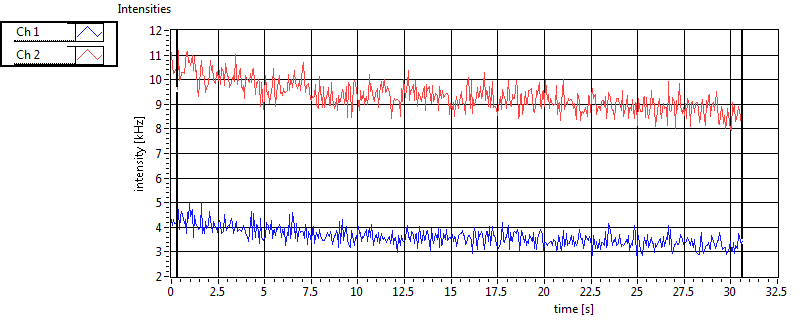
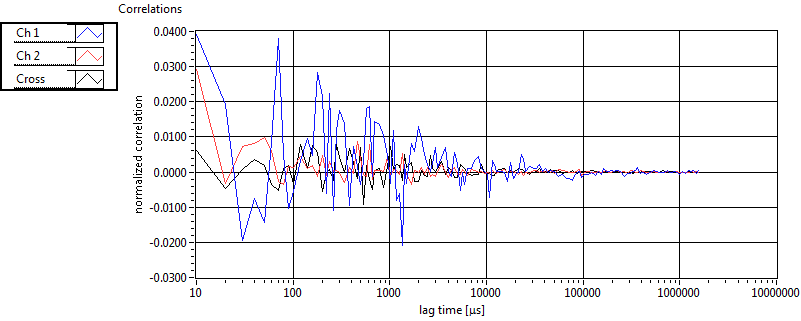
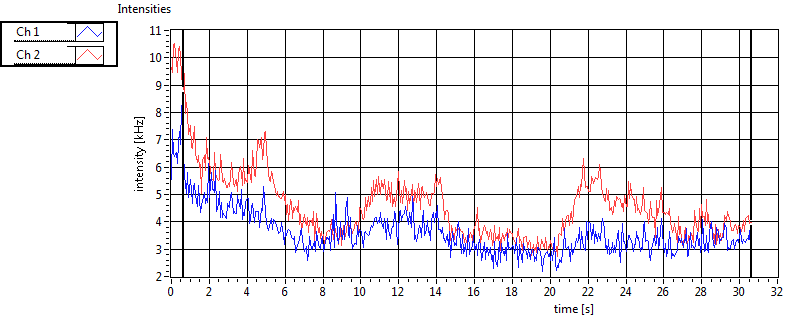
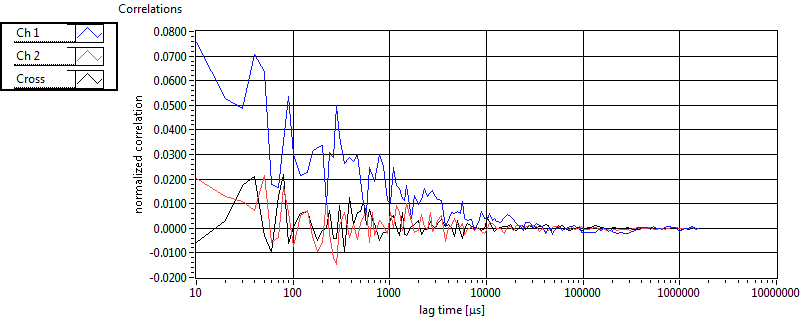
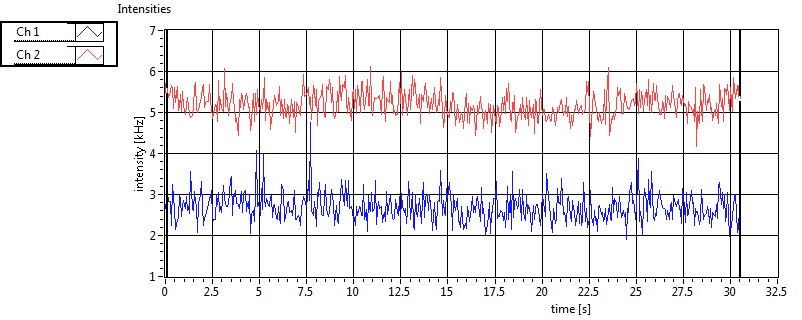
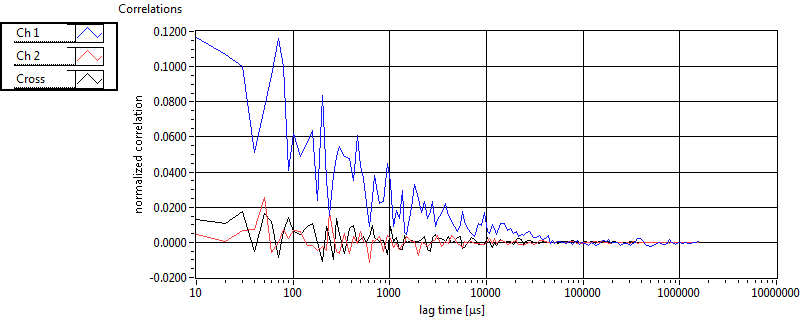
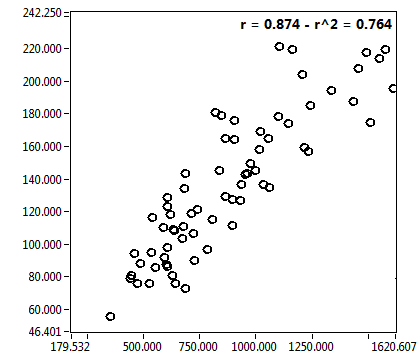
| 1 | 20120203\U01--V00--3155 |
|---|---|
| 2 | 20120228\U00--V00--3155 |
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 1.303E+2 | 6.691E+1 | 1.036E+3 | 4.640E+2 | 1.505E+1 | 1.139E+1 | 3.696E+1 | 5.617E+1 | 3.695E+2 | 2.893E+2 | 9.091E+1 | 3.177E+1 |
| N_corr | 1.638E+2 | 8.994E+1 | 1.602E+3 | 8.675E+2 | 2.336E+1 | 1.645E+1 | 3.959E+1 | 6.995E+1 | 5.589E+2 | 5.470E+2 | 1.689E+2 | 6.904E+1 |
| c [nM] | 8.107E+2 | 4.161E+2 | 4.804E+3 | 2.152E+3 | 7.735E+1 | 5.858E+1 | 2.299E+2 | 3.493E+2 | 1.714E+3 | 1.342E+3 | 4.674E+2 | 1.633E+2 |
| c_corr [nM] | 1.019E+3 | 5.594E+2 | 7.432E+3 | 4.024E+3 | 1.201E+2 | 8.459E+1 | 2.462E+2 | 4.351E+2 | 2.593E+3 | 2.537E+3 | 8.681E+2 | 3.549E+2 |
| ratioG | 1.002E-1 | 4.943E-2 | 4.984E-1 | 1.755E-2 | ||||||||
| ratioG_corr | 4.337E-2 | 4.714E-2 | 5.725E-1 | 1.698E-2 | ||||||||
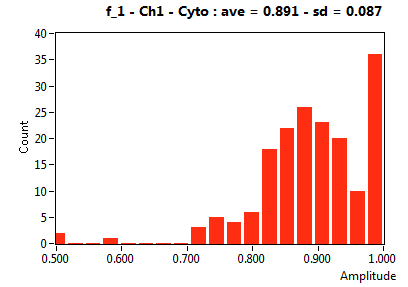
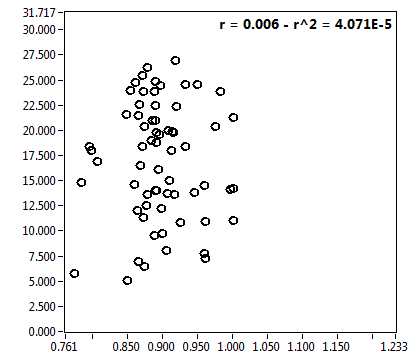
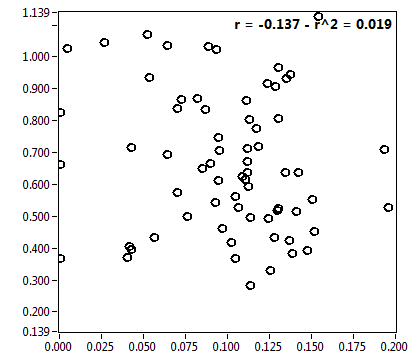
| f_1 | 5.755E-1 | 1.363E-1 | 3.964E-1 | 2.044E-1 | 8.397E-1 | 1.662E-1 | 8.907E-1 | 8.667E-2 | 7.062E-1 | 3.500E-1 | 9.391E-1 | 4.995E-2 |
| tauD_1 [ms] | 3.142E+0 | 3.002E+0 | 1.860E+0 | 2.220E-16 | 7.358E+0 | 1.089E+1 | 8.143E-1 | 6.639E-1 | 1.860E+0 | 2.220E-16 | 2.814E+0 | 2.990E-1 |
| D_1 [mum^2/s] | 6.771E+0 | 5.577E+0 | 6.926E+0 | 8.882E-16 | 1.644E+1 | 3.240E+1 | 2.019E+1 | 1.432E+1 | 6.926E+0 | 0.000E+0 | 4.309E+0 | 4.482E-1 |
| alpha_1 | 9.210E-1 | 1.051E-1 | 9.200E-1 | 2.220E-16 | 7.470E-1 | 2.034E-1 | 9.677E-1 | 1.372E-1 | 9.200E-1 | 1.110E-16 | 1.200E+0 | 0.000E+0 |
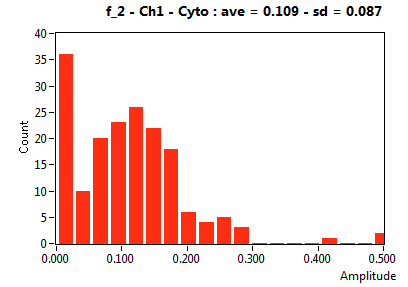
| f_2 | 4.245E-1 | 1.363E-1 | 6.036E-1 | 2.044E-1 | 1.603E-1 | 1.662E-1 | 1.093E-1 | 8.667E-2 | 2.938E-1 | 3.500E-1 | 6.094E-2 | 4.995E-2 |
| tauD_2 [ms] | 5.063E+1 | 3.686E+1 | 1.097E+2 | 1.231E+2 | 3.173E+2 | 7.743E+2 | 2.327E+1 | 1.824E+1 | 6.674E+1 | 7.170E+1 | 7.173E+1 | 1.155E+1 |
| D_2 [mum^2/s] | 2.918E-1 | 1.403E-1 | 3.759E-1 | 5.480E-1 | 9.863E-2 | 8.885E-2 | 6.590E-1 | 3.468E-1 | 1.137E+0 | 1.641E+0 | 1.714E-1 | 2.674E-2 |
| alpha_2 | 1.550E+0 | 2.601E-1 | 1.303E+0 | 3.019E-1 | 1.308E+0 | 3.478E-1 | 1.118E+0 | 1.614E-1 | 1.406E+0 | 3.996E-1 | 1.121E+0 | 5.288E-1 |
| tauD_m [ms] | 2.089E+1 | 1.137E+1 | 6.575E+1 | 7.324E+1 | 5.923E+1 | 1.259E+2 | 3.657E+0 | 4.388E+0 | 1.451E+1 | 2.431E+1 | 7.579E+0 | 4.327E+0 |
| D_m [mum^2/s] | 3.531E+0 | 2.367E+0 | 2.994E+0 | 1.311E+0 | 1.280E+1 | 2.611E+1 | 1.826E+1 | 1.390E+1 | 5.709E+0 | 1.227E+0 | 4.065E+0 | 5.600E-1 |
| R [1/s] | 1.111E+1 | 1.112E+1 | 6.050E+0 | 3.264E+0 | ||||||||
| CPM [kHz] | 9.303E-1 | 2.486E-1 | 4.253E-1 | 3.673E-1 | 4.639E-1 | 3.037E-1 | 2.317E-1 | 1.257E-1 | ||||
| CPM_corr [kHz] | 9.477E-1 | 2.516E-1 | 4.811E-1 | 4.149E-1 | 7.203E-1 | 4.922E-1 | 7.081E-1 | 1.734E+0 | ||||
| Int [kHz] | 1.173E+2 | 5.062E+1 | 1.721E+2 | 4.710E+1 | 1.296E+1 | 2.138E+1 | 6.462E+1 | 4.155E+1 | ||||
| Int_corr [kHz] | 1.495E+2 | 6.876E+1 | 2.939E+2 | 1.031E+2 | 1.677E+1 | 3.106E+1 | 1.090E+2 | 8.612E+1 | ||||
| # samples | 244 | 244 | 165 | 165 | 39 | 39 | 176 | 176 | 18 | 18 | 3 | 3 |
| # cells | 88 |
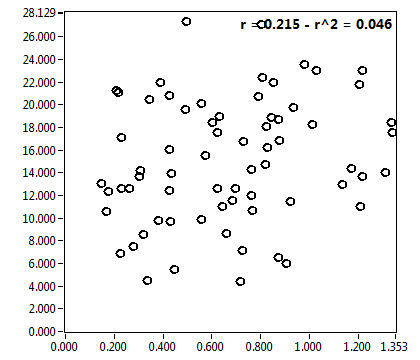
![c_corr [nM] - Ch1 - Nuc : ave = 1018.585 - sd = 559.369](3155_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 149.540 - sd = 68.759](3155_Int_corr_1n.png)
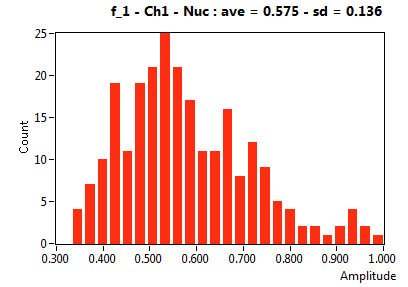
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 6.771 - sd = 5.577](3155_D_1_1n.png)
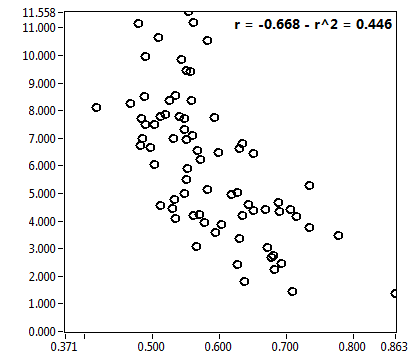
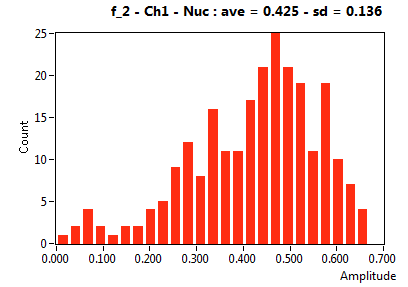
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.292 - sd = 0.140](3155_D_2_1n.png)
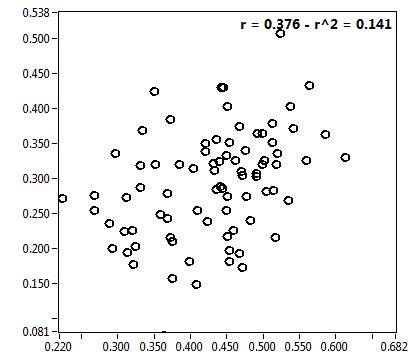
![CPM_corr [kHz] - Ch1 - Nuc : ave = 0.948 - sd = 0.252](3155_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 3.531 - sd = 2.367](3155_D_m_1n.png)
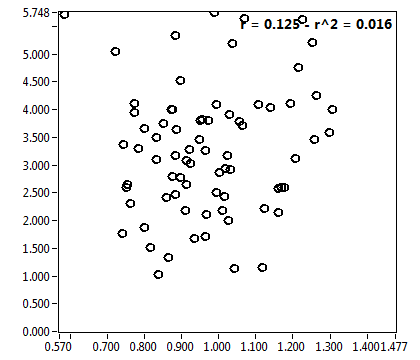
![c_corr [nM] - Ch1 - Nuc : ave = 1018.585 - sd = 559.369](3155_c_corr_1n.png)
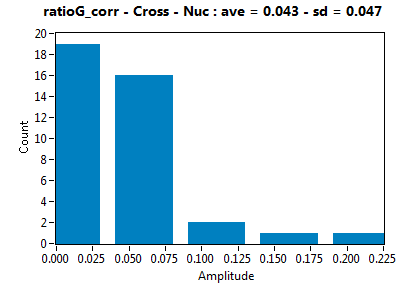
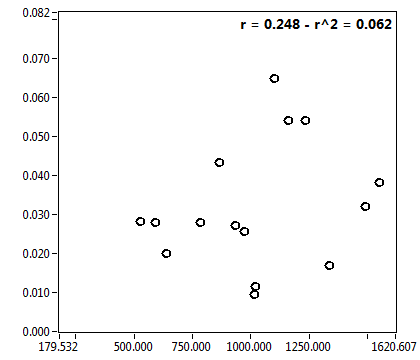
![c_corr [nM] - Ch1 - Cyto : ave = 246.190 - sd = 435.065](3155_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 16.772 - sd = 31.057](3155_Int_corr_1c.png)
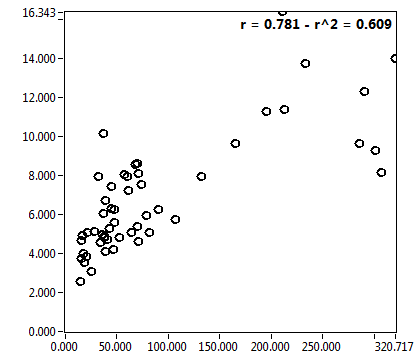
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 20.187 - sd = 14.324](3155_D_1_1c.png)
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 0.659 - sd = 0.347](3155_D_2_1c.png)
![CPM_corr [kHz] - Ch1 - Cyto : ave = 0.720 - sd = 0.492](3155_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 18.262 - sd = 13.897](3155_D_m_1c.png)