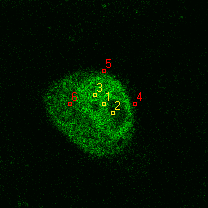
 |  |
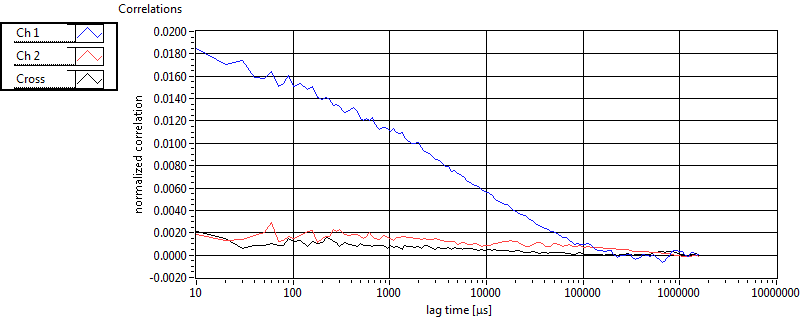
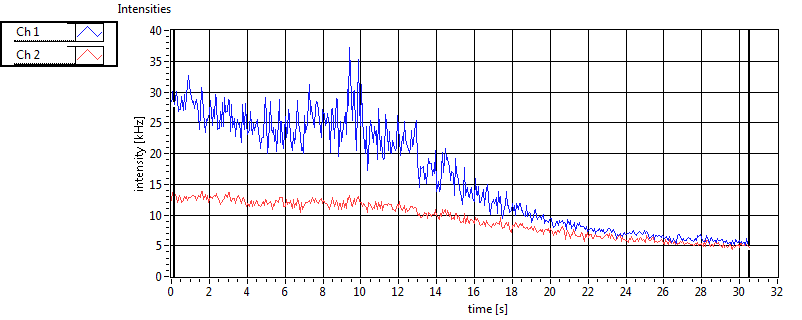
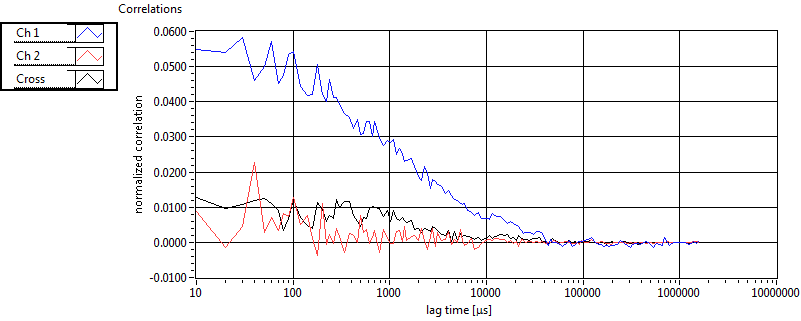
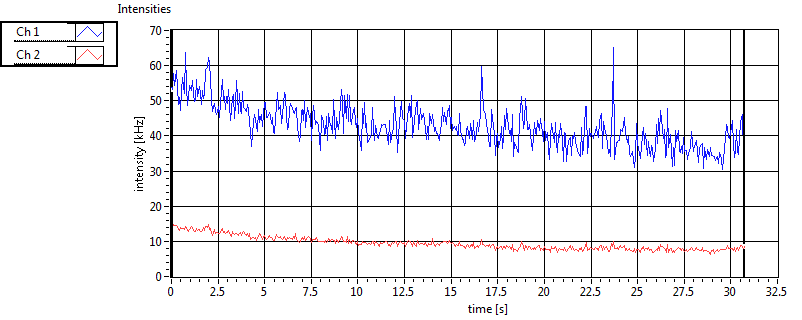
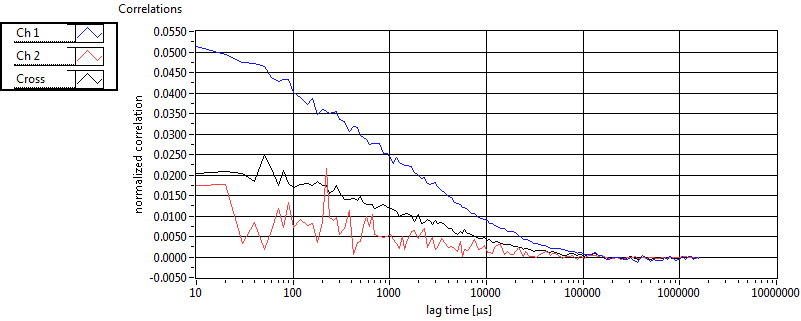
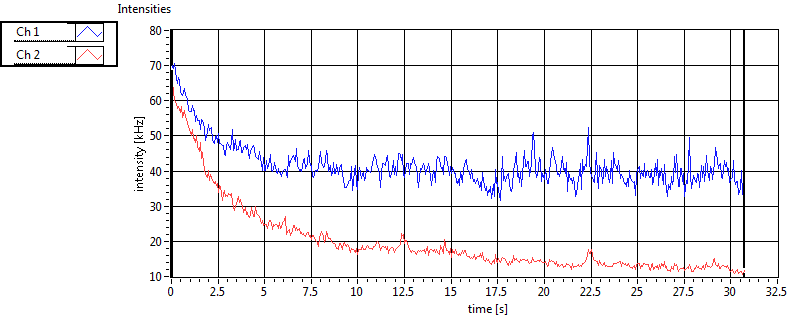
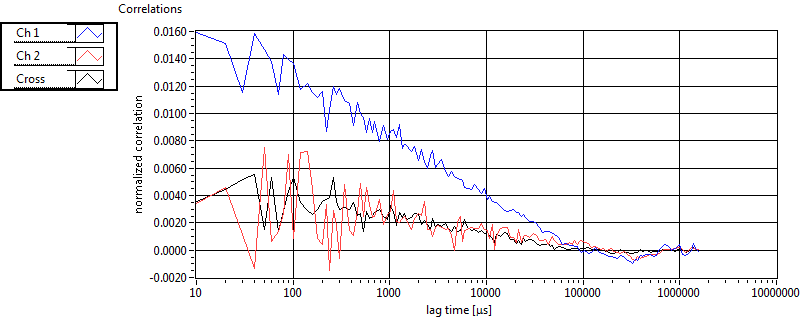
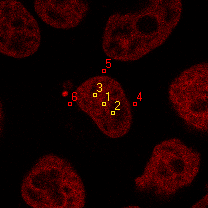
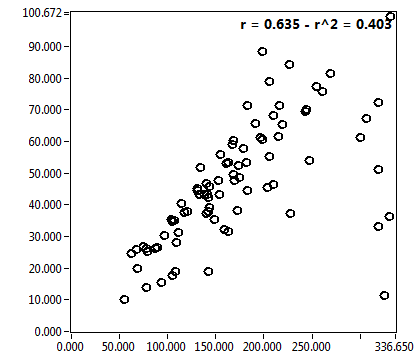
| 1 | 20110913\U02--V00--3140 |
|---|---|
| 2 | 20110914\U02--V01--3140 |
| 3 | 20110923\U03--V00--3140 |
| 4 | 20111011\U03--V01--3140 |
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 3.401E+1 | 2.505E+1 | 8.198E+2 | 3.311E+2 | 2.493E+0 | 4.115E+0 | 1.986E+1 | 1.149E+1 | 1.526E+2 | 1.847E+2 | 8.568E+0 | 7.266E+0 |
| N_corr | 3.862E+1 | 3.313E+1 | 1.223E+3 | 6.060E+2 | 3.525E+0 | 6.789E+0 | 2.344E+1 | 1.730E+1 | 1.783E+2 | 2.970E+2 | 1.160E+1 | 9.714E+0 |
| c [nM] | 2.115E+2 | 1.558E+2 | 3.803E+3 | 1.536E+3 | 1.282E+1 | 2.115E+1 | 1.235E+2 | 7.147E+1 | 7.077E+2 | 8.566E+2 | 4.405E+1 | 3.735E+1 |
| c_corr [nM] | 2.402E+2 | 2.061E+2 | 5.673E+3 | 2.811E+3 | 1.812E+1 | 3.490E+1 | 1.458E+2 | 1.076E+2 | 8.271E+2 | 1.377E+3 | 5.961E+1 | 4.994E+1 |
| ratioG | 4.912E-2 | 3.272E-2 | 4.004E-1 | 2.449E-1 | ||||||||
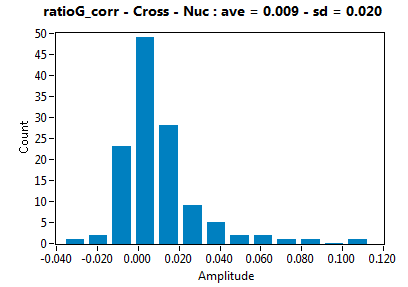
| ratioG_corr | 9.382E-3 | 2.001E-2 | 8.559E-2 | 7.853E-2 | ||||||||
| f_1 | 8.214E-1 | 1.016E-1 | 4.697E-1 | 2.235E-1 | 9.633E-1 | 9.614E-2 | 8.345E-1 | 1.150E-1 | 9.669E-1 | 1.127E-1 | 9.433E-1 | 7.118E-2 |
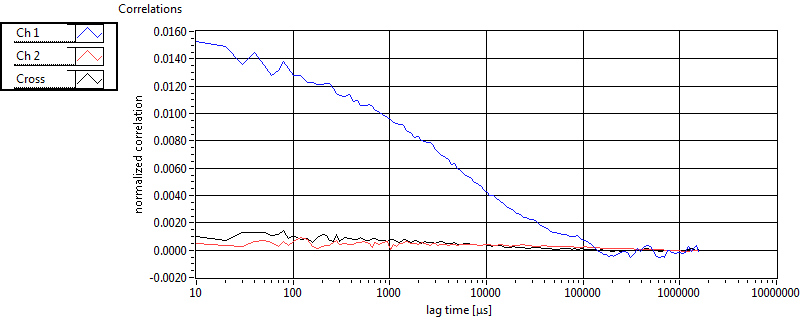
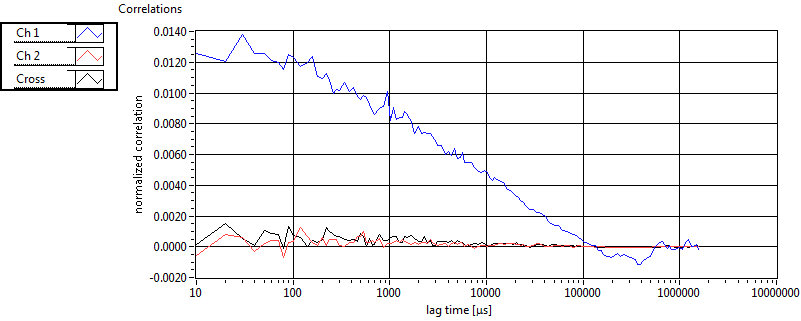
| tauD_1 [ms] | 1.373E+0 | 1.088E+0 | 1.877E+0 | 3.116E-2 | 1.479E+0 | 1.493E+0 | 1.005E+0 | 1.404E+0 | 4.564E+0 | 1.057E+1 | 7.763E-1 | 9.512E-1 |
| D_1 [mum^2/s] | 1.478E+1 | 1.225E+1 | 6.865E+0 | 1.165E-1 | 2.422E+1 | 2.982E+1 | 2.131E+1 | 1.503E+1 | 1.964E+1 | 2.837E+1 | 4.453E+1 | 3.517E+1 |
| alpha_1 | 9.838E-1 | 1.289E-1 | 9.658E-1 | 3.958E-2 | 8.572E-1 | 2.330E-1 | 9.583E-1 | 1.880E-1 | 8.564E-1 | 2.421E-1 | 8.294E-1 | 2.227E-1 |
| f_2 | 1.786E-1 | 1.016E-1 | 5.303E-1 | 2.235E-1 | 3.667E-2 | 9.614E-2 | 1.655E-1 | 1.150E-1 | 3.311E-2 | 1.127E-1 | 5.671E-2 | 7.118E-2 |
| tauD_2 [ms] | 2.414E+1 | 1.543E+1 | 1.023E+2 | 1.302E+2 | 9.488E+1 | 7.644E+1 | 2.902E+1 | 2.728E+1 | 2.221E+1 | 5.495E+1 | 4.092E+1 | 3.519E+1 |
| D_2 [mum^2/s] | 6.719E-1 | 4.802E-1 | 5.906E-1 | 8.965E-1 | 1.826E-1 | 1.022E-1 | 6.204E-1 | 4.465E-1 | 2.171E+0 | 8.913E-1 | 5.359E-1 | 3.908E-1 |
| alpha_2 | 1.420E+0 | 3.457E-1 | 1.380E+0 | 3.556E-1 | 1.159E+0 | 2.280E-1 | 1.217E+0 | 2.976E-1 | 1.133E+0 | 1.472E-1 | 1.166E+0 | 2.183E-1 |
| tauD_m [ms] | 5.983E+0 | 4.864E+0 | 5.405E+1 | 8.056E+1 | 7.059E+0 | 2.145E+1 | 6.705E+0 | 8.642E+0 | 7.955E+0 | 1.693E+1 | 3.102E+0 | 5.601E+0 |
| D_m [mum^2/s] | 1.273E+1 | 1.147E+1 | 3.639E+0 | 1.207E+0 | 2.362E+1 | 2.965E+1 | 1.860E+1 | 1.431E+1 | 1.942E+1 | 2.848E+1 | 4.231E+1 | 3.402E+1 |
| R [1/s] | 4.844E+1 | 5.529E+1 | 3.423E+0 | 9.646E+0 | ||||||||
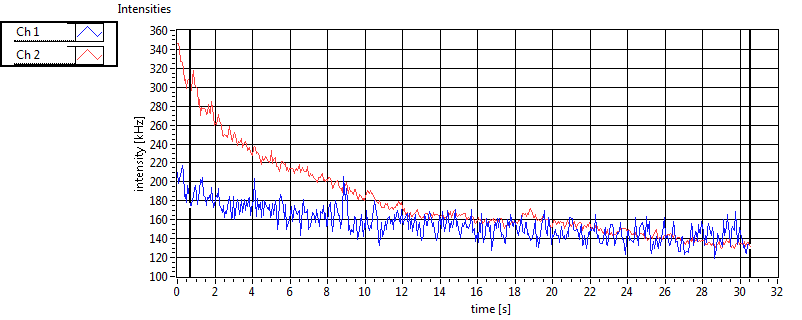
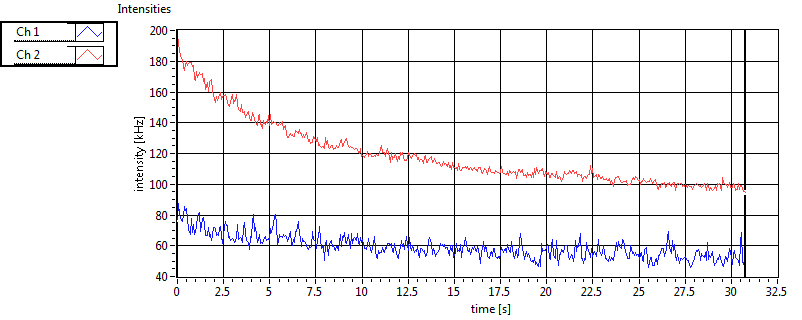
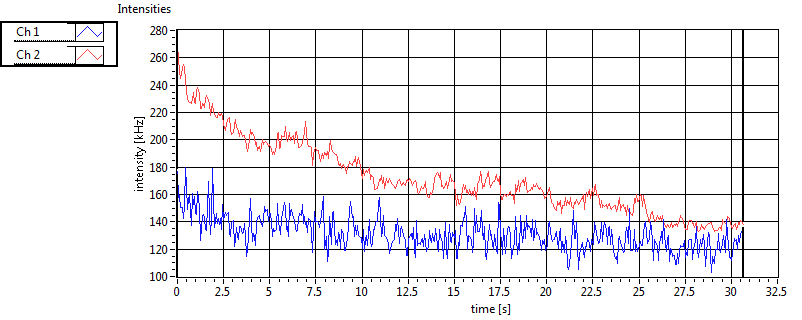
| CPM [kHz] | 1.551E+0 | 4.596E-1 | 3.335E-1 | 3.353E-1 | 1.410E+0 | 4.806E-1 | 2.098E-1 | 1.726E-1 | ||||
| CPM_corr [kHz] | 1.627E+0 | 4.631E-1 | 3.618E-1 | 3.651E-1 | 1.787E+0 | 3.254E+0 | 7.643E-1 | 7.599E-1 | ||||
| Int [kHz] | 5.773E+1 | 4.572E+1 | 1.451E+2 | 4.234E+1 | 3.100E+1 | 1.981E+1 | 3.138E+1 | 4.263E+1 | ||||
| Int_corr [kHz] | 6.700E+1 | 5.685E+1 | 2.275E+2 | 7.814E+1 | 3.783E+1 | 2.393E+1 | 4.673E+1 | 6.609E+1 | ||||
| # samples | 197 | 197 | 145 | 145 | 124 | 124 | 309 | 309 | 89 | 89 | 81 | 81 |
| # cells | 107 |
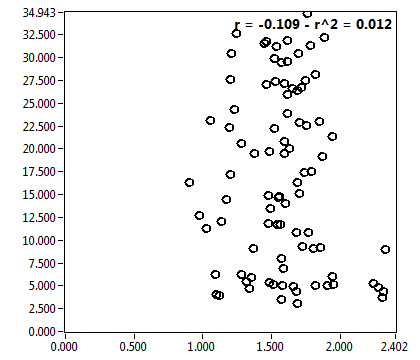
![c_corr [nM] - Ch1 - Nuc : ave = 240.169 - sd = 206.072](3140_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 67.000 - sd = 56.846](3140_Int_corr_1n.png)
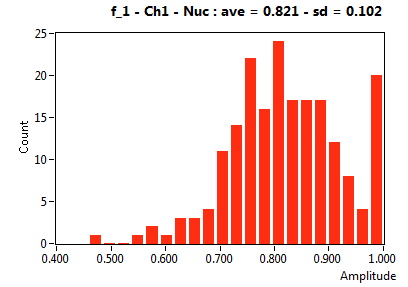
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 14.776 - sd = 12.245](3140_D_1_1n.png)
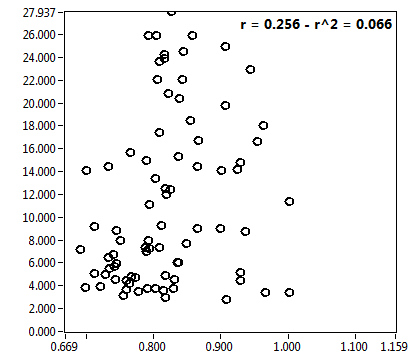
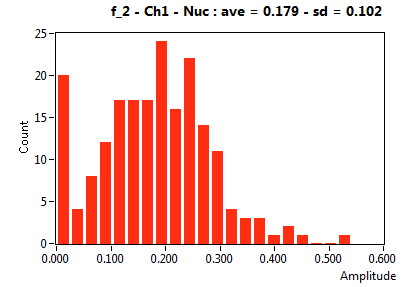
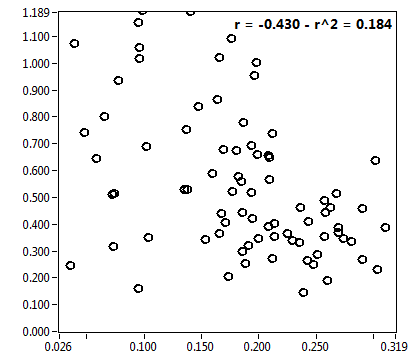
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.672 - sd = 0.480](3140_D_2_1n.png)
![CPM_corr [kHz] - Ch1 - Nuc : ave = 1.627 - sd = 0.463](3140_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 12.729 - sd = 11.470](3140_D_m_1n.png)
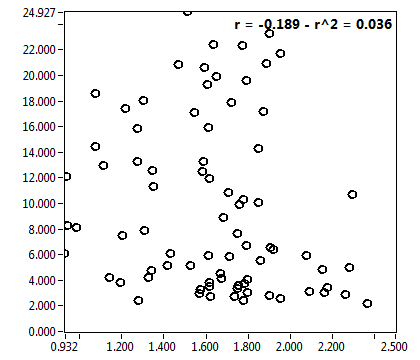
![c_corr [nM] - Ch1 - Nuc : ave = 240.169 - sd = 206.072](3140_c_corr_1n.png)
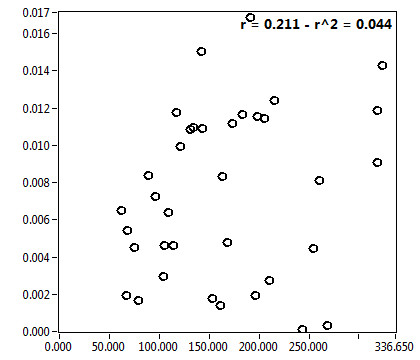
![c_corr [nM] - Ch1 - Cyto : ave = 145.756 - sd = 107.602](3140_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 37.826 - sd = 23.927](3140_Int_corr_1c.png)
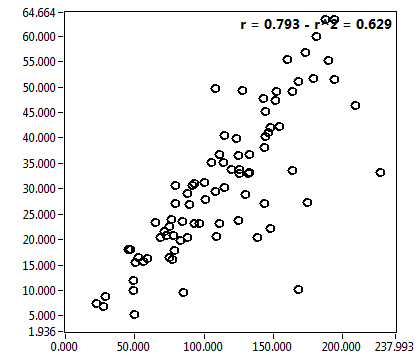
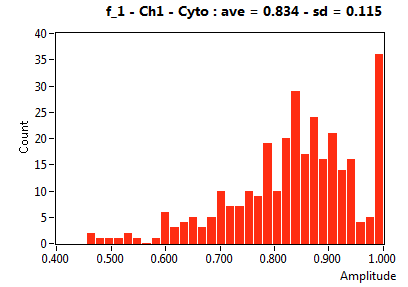
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 21.310 - sd = 15.027](3140_D_1_1c.png)
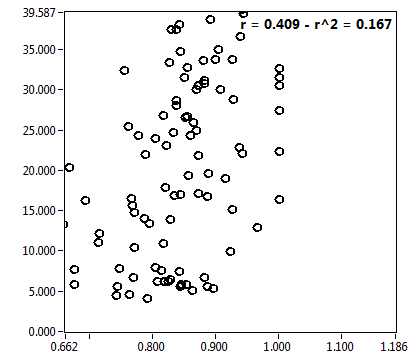
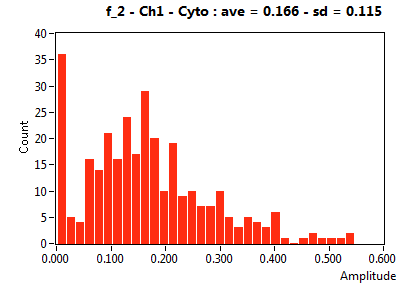
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 0.620 - sd = 0.447](3140_D_2_1c.png)
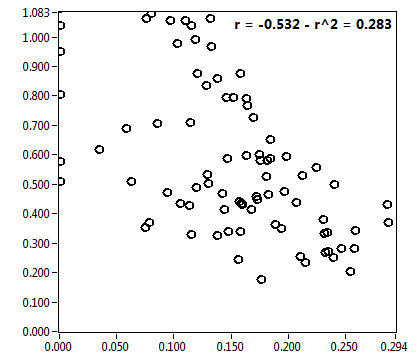
![CPM_corr [kHz] - Ch1 - Cyto : ave = 1.787 - sd = 3.254](3140_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 18.595 - sd = 14.313](3140_D_m_1c.png)