 |  |
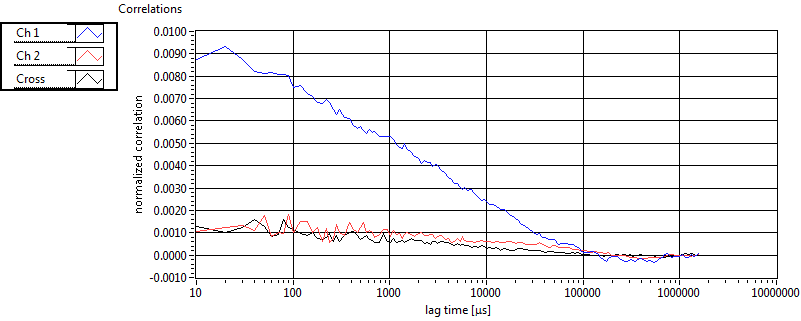
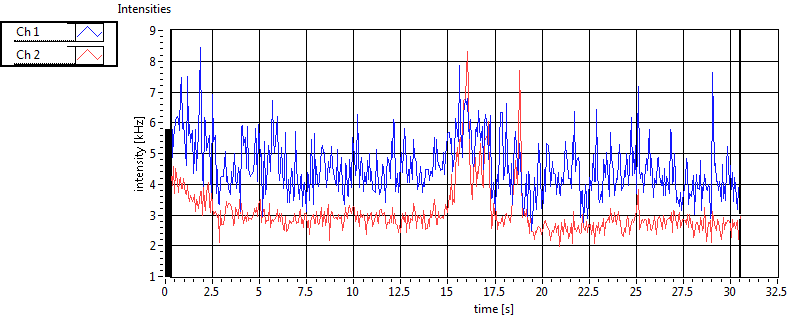
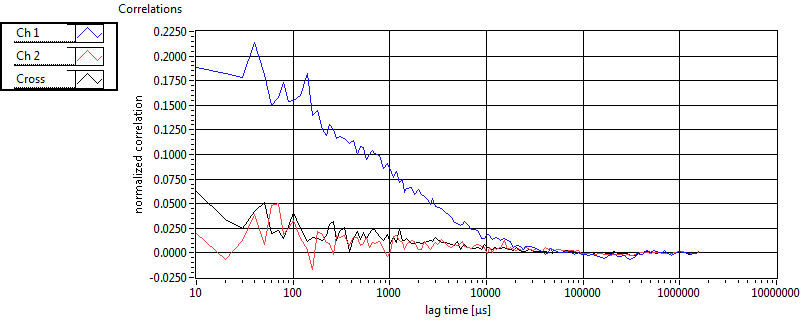
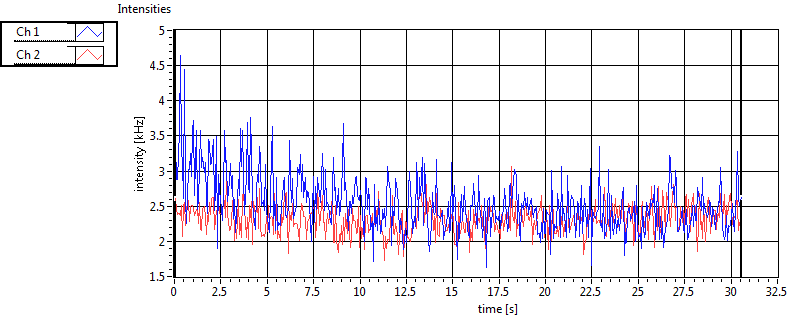
| 1 | 20110915\U03--V01--2256 |
|---|---|
| 2 | 20110916\U03--V01--2256 |
| 3 | 20110922\U00--V01--2256 |
| 4 | 20110930\U02--V01--2256 |
| 5 | 20111011\U00--V01--2256 |
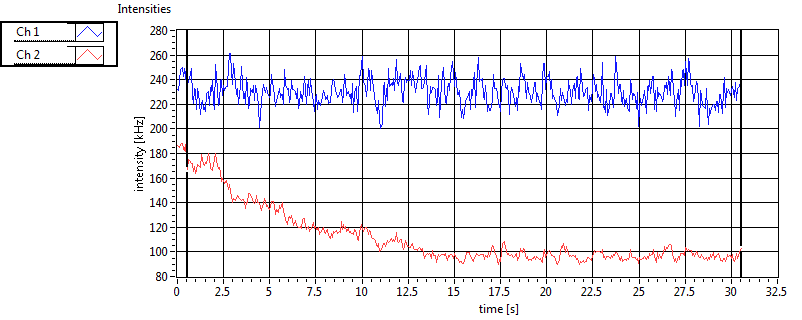
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 1.192E+2 | 9.285E+1 | 8.667E+2 | 4.005E+2 | 2.297E+1 | 3.017E+1 | 2.641E+1 | 3.535E+1 | 3.647E+2 | 1.416E+2 | 2.759E+1 | 2.823E+1 |
| N_corr | 1.368E+2 | 1.113E+2 | 1.151E+3 | 6.680E+2 | 3.046E+1 | 4.316E+1 | 3.244E+1 | 6.417E+1 | 5.694E+2 | 3.497E+2 | 3.047E+1 | 2.997E+1 |
| c [nM] | 7.413E+2 | 5.775E+2 | 4.020E+3 | 1.857E+3 | 1.181E+2 | 1.551E+2 | 1.642E+2 | 2.199E+2 | 1.692E+3 | 6.566E+2 | 1.418E+2 | 1.451E+2 |
| c_corr [nM] | 8.507E+2 | 6.921E+2 | 5.340E+3 | 3.098E+3 | 1.566E+2 | 2.219E+2 | 2.017E+2 | 3.991E+2 | 2.641E+3 | 1.622E+3 | 1.567E+2 | 1.541E+2 |
| ratioG | 1.260E-1 | 9.941E-2 | 2.552E-1 | 1.995E-1 | ||||||||
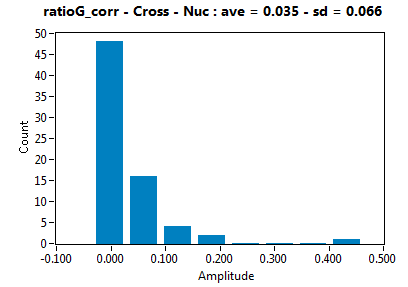
| ratioG_corr | 3.481E-2 | 6.591E-2 | 2.179E-1 | 2.240E-1 | ||||||||
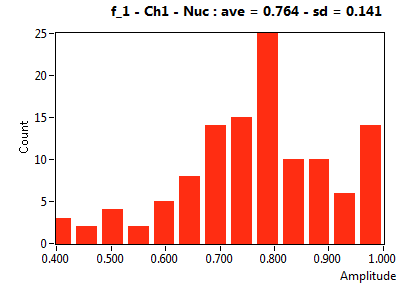
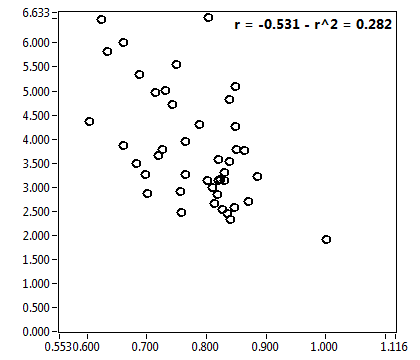
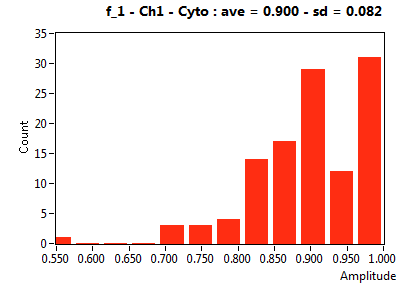
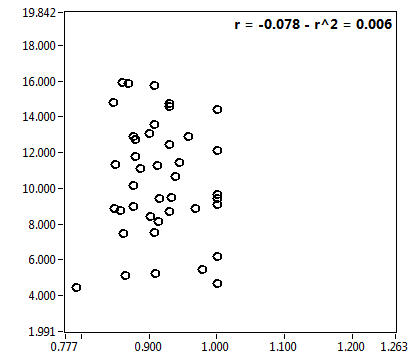
| f_1 | 7.644E-1 | 1.406E-1 | 4.644E-1 | 1.969E-1 | 8.999E-1 | 1.330E-1 | 9.002E-1 | 8.222E-2 | 6.000E-1 | 3.294E-1 | 9.297E-1 | 1.003E-1 |
| tauD_1 [ms] | 3.353E+0 | 2.047E+0 | 1.809E+0 | 2.181E-2 | 2.862E+0 | 2.046E+0 | 1.174E+0 | 8.001E-1 | 1.814E+0 | 2.528E-2 | 2.265E+0 | 1.096E+0 |
| D_1 [mum^2/s] | 4.399E+0 | 3.127E+0 | 7.121E+0 | 8.391E-2 | 6.381E+0 | 5.391E+0 | 1.246E+1 | 6.979E+0 | 7.104E+0 | 9.727E-2 | 6.653E+0 | 3.383E+0 |
| alpha_1 | 9.396E-1 | 1.085E-1 | 9.200E-1 | 1.110E-16 | 7.681E-1 | 2.132E-1 | 9.367E-1 | 1.240E-1 | 9.200E-1 | 0.000E+0 | 9.999E-1 | 2.127E-1 |
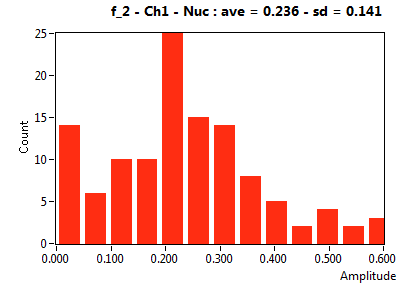
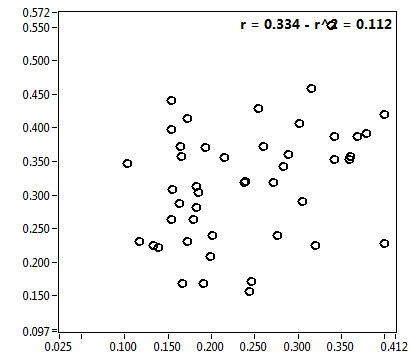
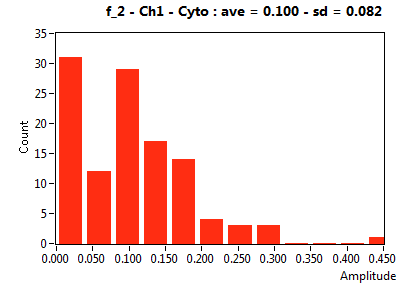
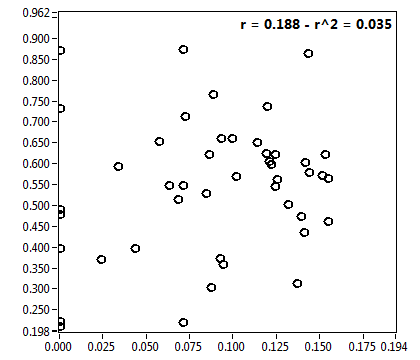
| f_2 | 2.356E-1 | 1.406E-1 | 5.356E-1 | 1.969E-1 | 1.001E-1 | 1.330E-1 | 9.981E-2 | 8.222E-2 | 4.000E-1 | 3.294E-1 | 7.027E-2 | 1.003E-1 |
| tauD_2 [ms] | 4.578E+1 | 3.454E+1 | 1.130E+2 | 1.103E+2 | 1.215E+2 | 9.721E+1 | 2.458E+1 | 1.569E+1 | 7.676E+1 | 9.001E+1 | 7.015E+1 | 3.140E+1 |
| D_2 [mum^2/s] | 3.174E-1 | 1.470E-1 | 4.947E-1 | 7.841E-1 | 1.461E-1 | 8.079E-2 | 5.559E-1 | 2.385E-1 | 1.106E+0 | 1.465E+0 | 2.162E-1 | 1.086E-1 |
| alpha_2 | 1.500E+0 | 3.472E-1 | 1.254E+0 | 3.149E-1 | 1.256E+0 | 3.005E-1 | 1.107E+0 | 2.178E-1 | 1.291E+0 | 4.015E-1 | 1.274E+0 | 3.544E-1 |
| tauD_m [ms] | 1.174E+1 | 7.280E+0 | 6.026E+1 | 6.385E+1 | 1.875E+1 | 3.410E+1 | 3.760E+0 | 4.505E+0 | 2.710E+1 | 4.214E+1 | 8.479E+0 | 9.232E+0 |
| D_m [mum^2/s] | 3.195E+0 | 1.744E+0 | 3.616E+0 | 1.191E+0 | 5.626E+0 | 4.529E+0 | 1.116E+1 | 5.845E+0 | 5.064E+0 | 1.438E+0 | 6.257E+0 | 3.475E+0 |
| R [1/s] | 1.293E+1 | 1.379E+1 | 7.615E+0 | 1.065E+1 | ||||||||
| CPM [kHz] | 1.458E+0 | 4.375E-1 | 3.608E-1 | 2.808E-1 | 6.123E-1 | 4.159E-1 | 2.706E-1 | 1.211E-1 | ||||
| CPM_corr [kHz] | 1.489E+0 | 4.403E-1 | 4.380E-1 | 3.454E-1 | 9.482E-1 | 5.918E-1 | 3.080E-1 | 1.368E-1 | ||||
| Int [kHz] | 1.724E+2 | 1.245E+2 | 1.553E+2 | 4.654E+1 | 1.384E+1 | 2.300E+1 | 8.153E+1 | 2.809E+1 | ||||
| Int_corr [kHz] | 1.995E+2 | 1.506E+2 | 2.373E+2 | 8.471E+1 | 2.097E+1 | 5.470E+1 | 1.472E+2 | 9.669E+1 | ||||
| # samples | 118 | 118 | 83 | 83 | 71 | 71 | 114 | 114 | 13 | 13 | 11 | 11 |
| # cells | 57 |
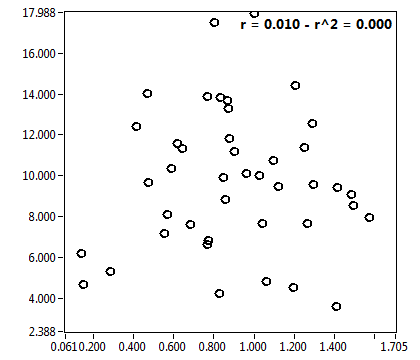
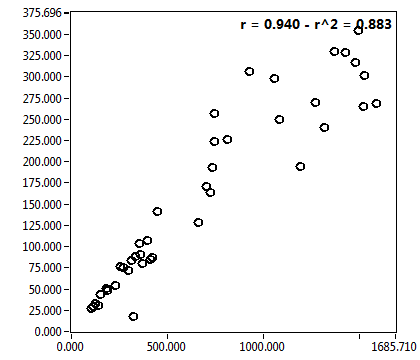
![c_corr [nM] - Ch1 - Nuc : ave = 850.671 - sd = 692.144](2256_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 199.471 - sd = 150.601](2256_Int_corr_1n.png)
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 4.399 - sd = 3.127](2256_D_1_1n.png)
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 0.317 - sd = 0.147](2256_D_2_1n.png)
![CPM_corr [kHz] - Ch1 - Nuc : ave = 1.489 - sd = 0.440](2256_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 3.195 - sd = 1.744](2256_D_m_1n.png)
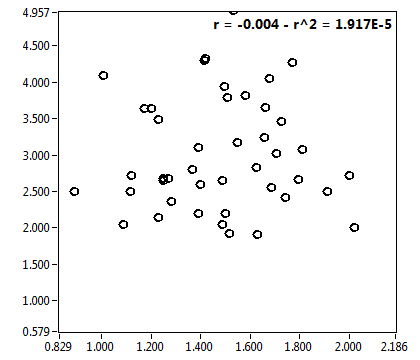
![c_corr [nM] - Ch1 - Nuc : ave = 850.671 - sd = 692.144](2256_c_corr_1n.png)
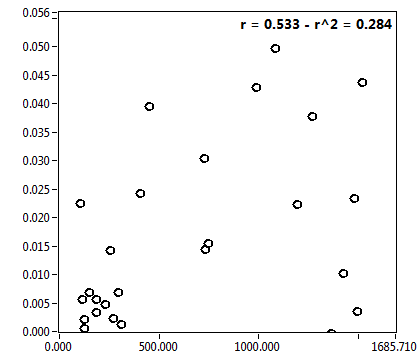
![c_corr [nM] - Ch1 - Cyto : ave = 201.727 - sd = 399.073](2256_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 20.967 - sd = 54.703](2256_Int_corr_1c.png)
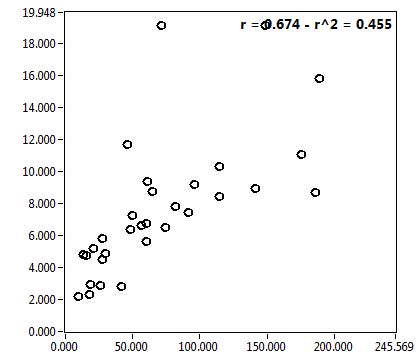
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 12.460 - sd = 6.979](2256_D_1_1c.png)
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 0.556 - sd = 0.239](2256_D_2_1c.png)
![CPM_corr [kHz] - Ch1 - Cyto : ave = 0.948 - sd = 0.592](2256_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 11.155 - sd = 5.845](2256_D_m_1c.png)