 |  |
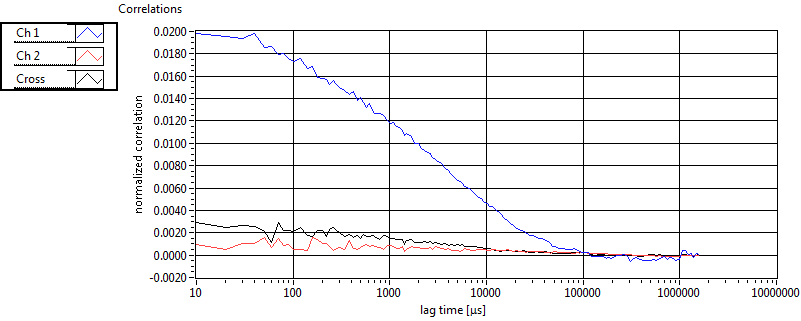
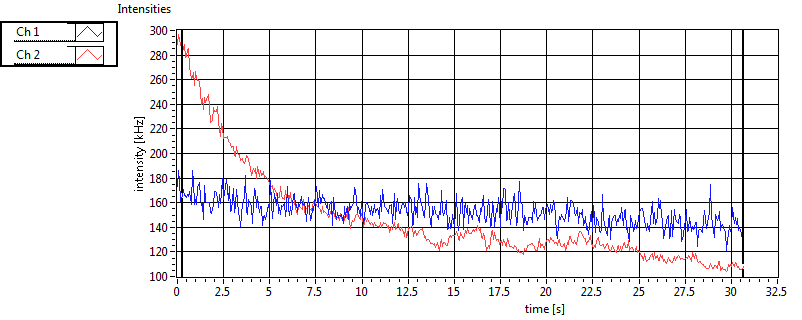
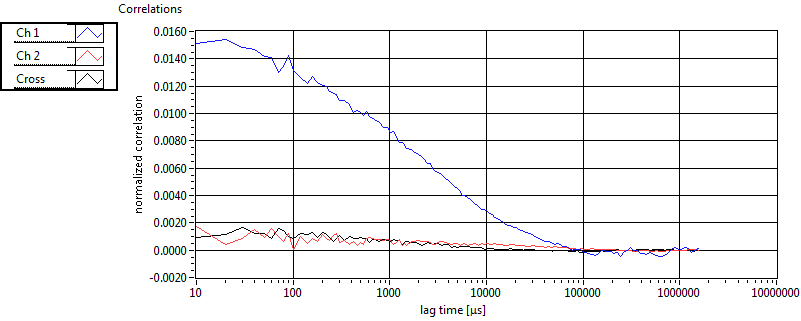
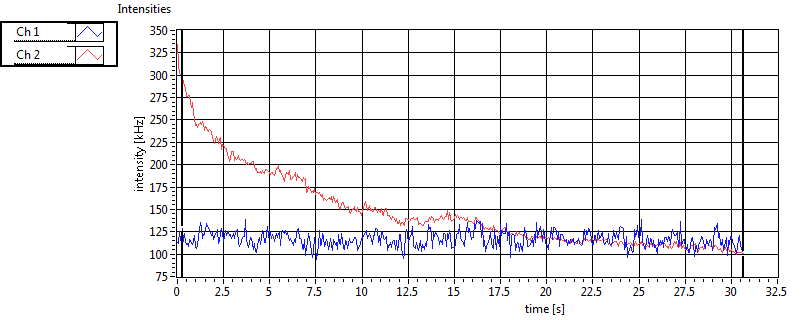
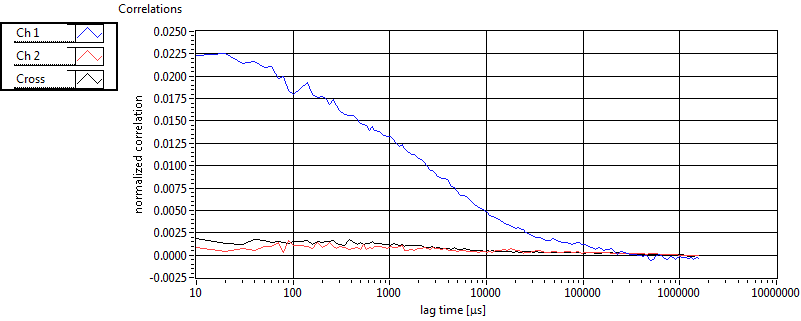
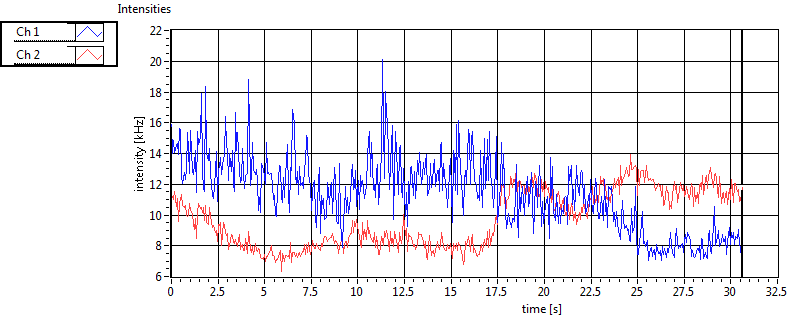
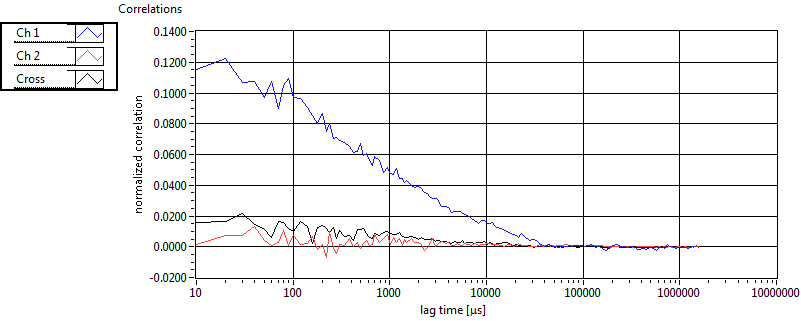
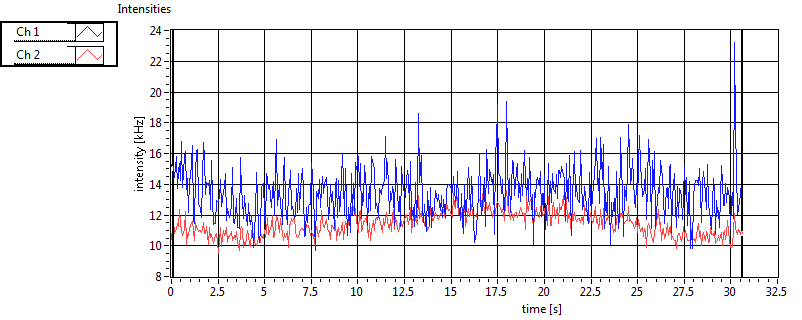
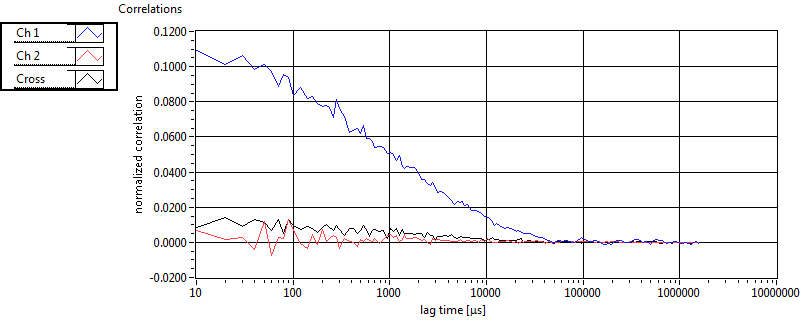
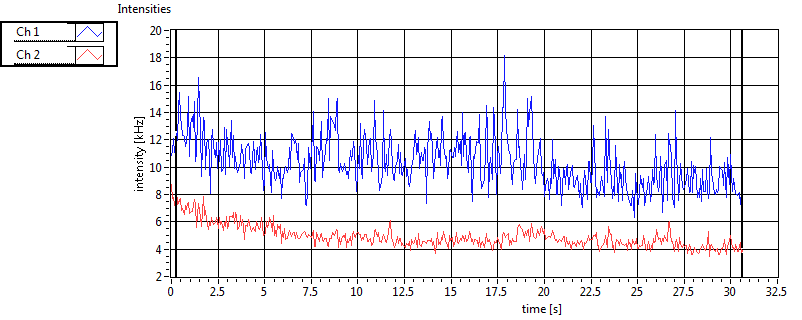
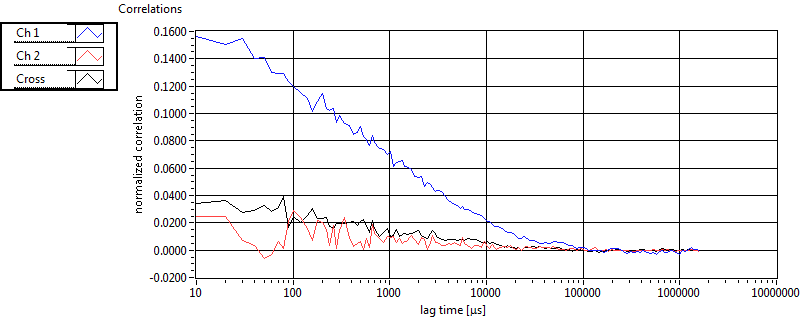
| 1 | 20111130\U00--V01--1153 |
|---|---|
| 2 | 20120201\U02--V00--1153 |
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 4.345E+1 | 1.637E+0 | 1.355E+3 | 1.794E+2 | 3.755E+1 | 4.964E+0 | ||||||
| N_corr | 3.921E+1 | 6.642E-1 | 2.107E+3 | 3.019E+2 | 3.991E+1 | 8.939E+0 | ||||||
| c [nM] | 2.702E+2 | 1.018E+1 | 6.287E+3 | 8.320E+2 | 2.335E+2 | 3.087E+1 | ||||||
| c_corr [nM] | 2.439E+2 | 4.131E+0 | 9.772E+3 | 1.401E+3 | 2.482E+2 | 5.560E+1 | ||||||
| ratioG | ||||||||||||
| ratioG_corr | ||||||||||||
| f_1 | 7.147E-1 | 5.920E-3 | 4.353E-1 | 1.176E-1 | 9.324E-1 | 5.004E-2 | ||||||
| tauD_1 [ms] | 3.902E-1 | 1.083E-2 | 1.860E+0 | 0.000E+0 | 2.265E-1 | 2.453E-2 | ||||||
| D_1 [mum^2/s] | 2.721E+1 | 7.554E-1 | 6.926E+0 | 0.000E+0 | 4.743E+1 | 5.540E+0 | ||||||
| alpha_1 | 1.100E+0 | 0.000E+0 | 9.200E-1 | 0.000E+0 | 1.014E+0 | 1.220E-1 | ||||||
| f_2 | 2.853E-1 | 5.920E-3 | 5.647E-1 | 1.176E-1 | 6.757E-2 | 5.004E-2 | ||||||
| tauD_2 [ms] | 5.017E+0 | 6.500E-4 | 2.640E+1 | 1.254E+1 | 5.002E+0 | 1.757E-3 | ||||||
| D_2 [mum^2/s] | 2.115E+0 | 2.740E-4 | 6.665E-1 | 3.943E-1 | 2.121E+0 | 7.449E-4 | ||||||
| alpha_2 | 1.032E+0 | 2.685E-2 | 1.530E+0 | 3.728E-1 | 9.101E-1 | 1.803E-1 | ||||||
| tauD_m [ms] | 1.710E+0 | 1.983E-2 | 1.426E+1 | 4.584E+0 | 5.497E-1 | 2.315E-1 | ||||||
| D_m [mum^2/s] | 2.005E+1 | 3.912E-1 | 3.437E+0 | 4.965E-1 | 4.428E+1 | 4.868E+0 | ||||||
| R [1/s] | ||||||||||||
| CPM [kHz] | 1.138E+0 | 2.230E-2 | 2.683E-1 | 3.785E-2 | 1.101E+0 | 1.679E-1 | ||||||
| CPM_corr [kHz] | 1.178E+0 | 2.238E-2 | 2.763E-1 | 3.835E-2 | 1.147E+0 | 1.764E-1 | ||||||
| Int [kHz] | 6.056E+1 | 2.490E-1 | 3.269E+2 | 3.605E+1 | 4.806E+1 | 3.615E-1 | ||||||
| Int_corr [kHz] | 5.662E+1 | 1.374E+0 | 5.279E+2 | 9.534E+1 | 5.252E+1 | 4.635E+0 | ||||||
| # samples | 2 | 2 | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 | 0 | 0 |
| # cells | 2 |
![c_corr [nM] - Ch1 - Nuc : ave = 243.867 - sd = 4.131](1153_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 56.617 - sd = 1.374](1153_Int_corr_1n.png)
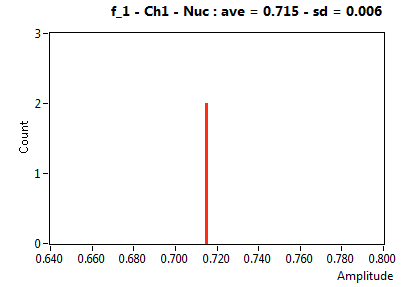
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 27.212 - sd = 0.755](1153_D_1_1n.png)
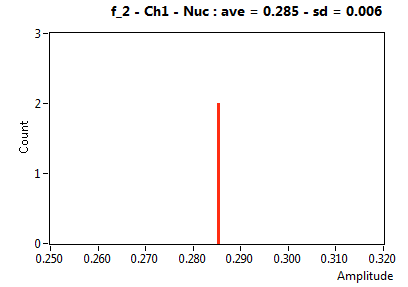
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 2.115 - sd = 0.000](1153_D_2_1n.png)
![CPM_corr [kHz] - Ch1 - Nuc : ave = 1.178 - sd = 0.022](1153_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 20.048 - sd = 0.391](1153_D_m_1n.png)
![c_corr [nM] - Ch1 - Nuc : ave = 243.867 - sd = 4.131](1153_c_corr_1n.png)
![c_corr [nM] - Ch1 - Cyto : ave = 248.240 - sd = 55.597](1153_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 52.520 - sd = 4.635](1153_Int_corr_1c.png)
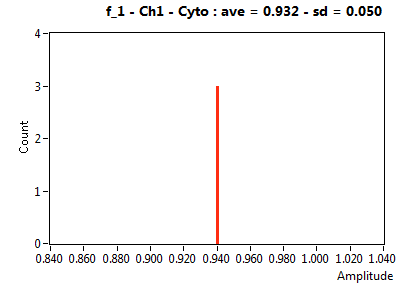
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 47.432 - sd = 5.540](1153_D_1_1c.png)
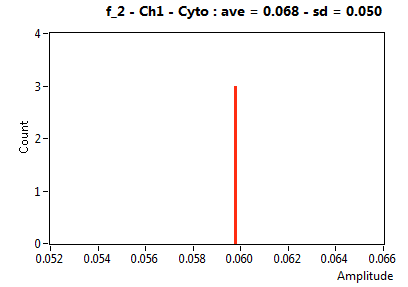
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 2.121 - sd = 0.001](1153_D_2_1c.png)
![CPM_corr [kHz] - Ch1 - Cyto : ave = 1.147 - sd = 0.176](1153_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 44.284 - sd = 4.868](1153_D_m_1c.png)