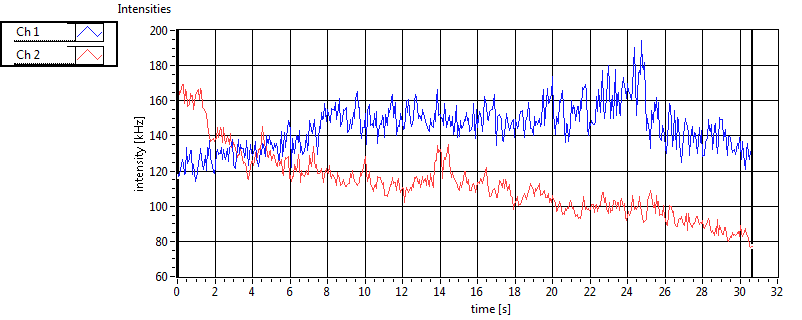
 |  |
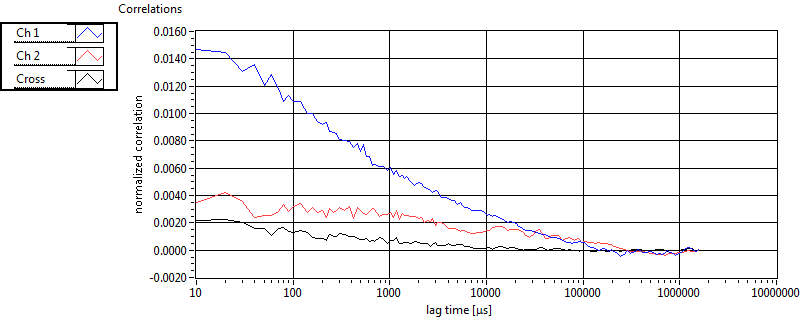
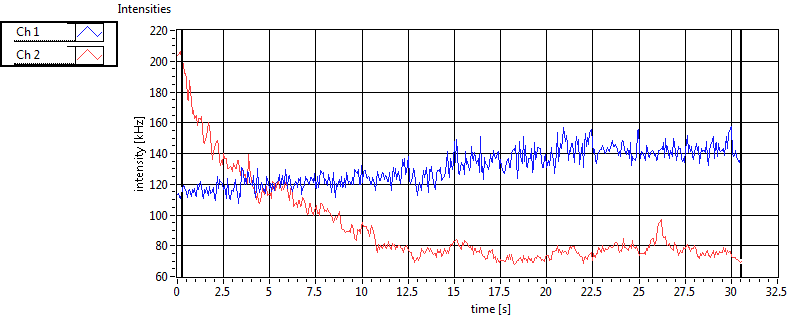
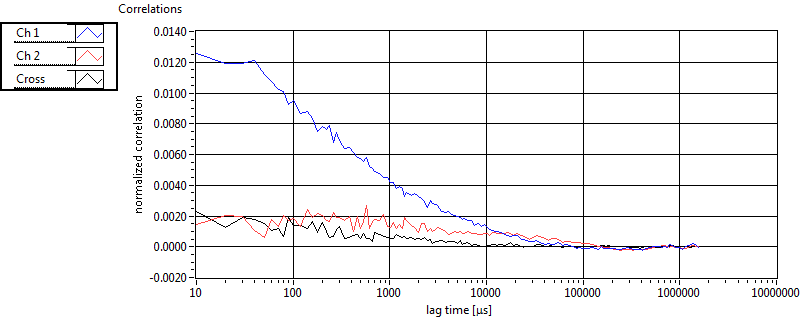
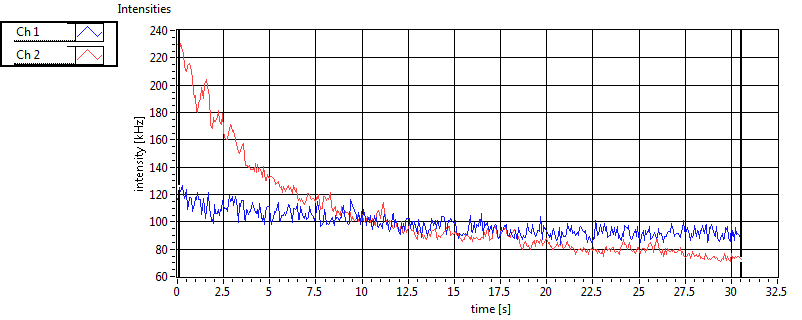
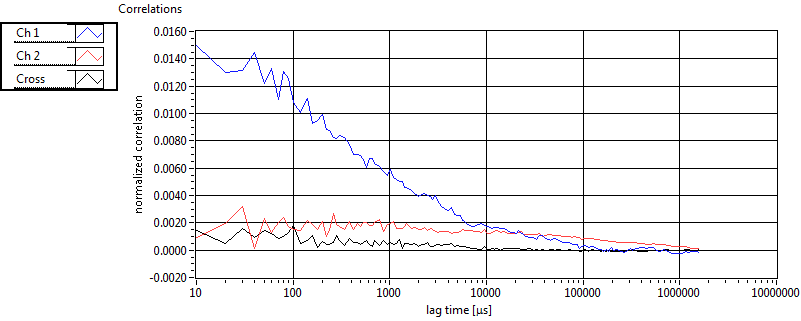
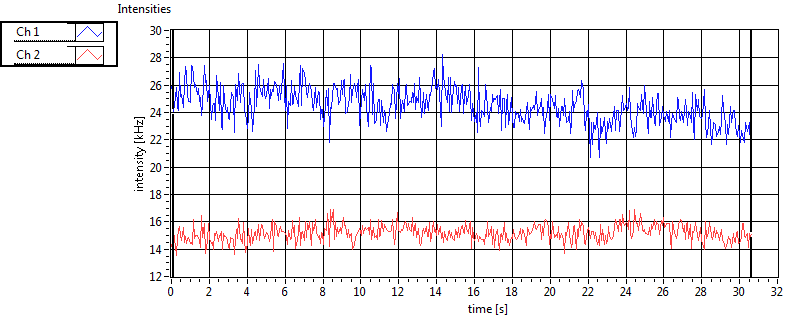
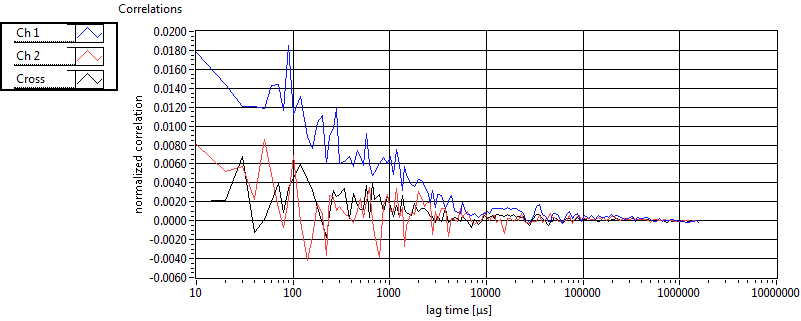
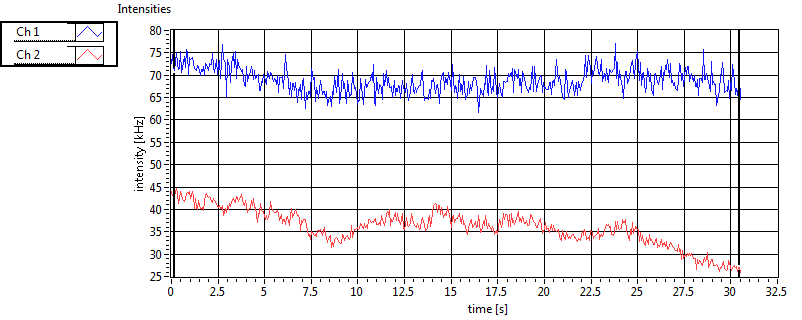
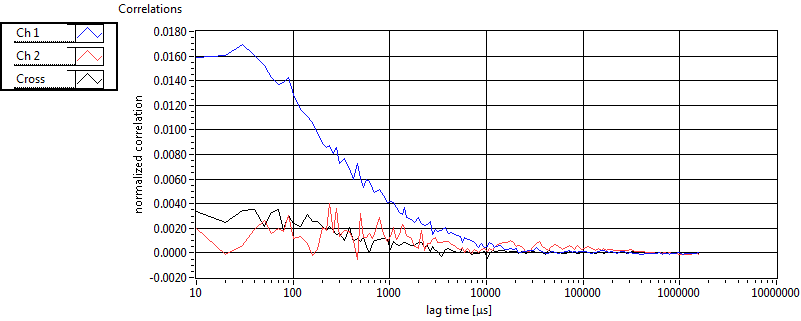
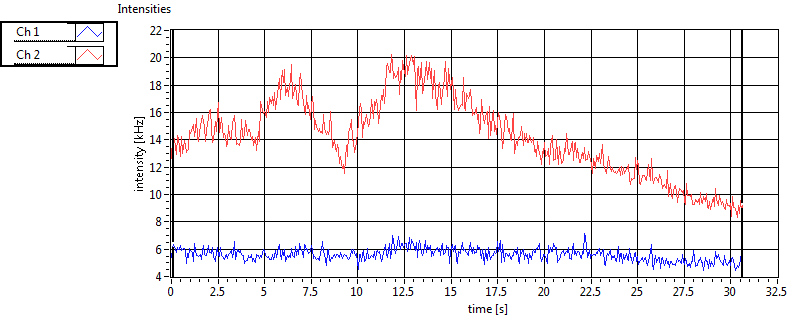
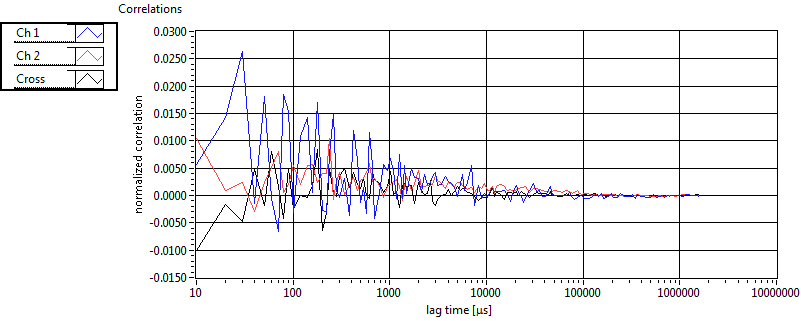
| 1 | 20110915\U01--V00--0592 |
|---|---|
| 2 | 20110916\U01--V00--0592 |
| 3 | 20110922\U01--V00--0592 |
| 4 | 20111011\U01--V00--0592 |
| Ch1 ave Nuc |
Ch1 sd Nuc |
Ch2 ave Nuc |
Ch2 sd Nuc |
Cross ave Nuc |
Cross sd Nuc |
Ch1 ave Cyto |
Ch1 sd Cyto |
Ch2 ave Cyto |
Ch2 sd Cyto |
Cross ave Cyto |
Cross sd Cyto |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 4.520E+1 | 2.753E+1 | 1.083E+3 | 4.696E+2 | 3.915E+0 | 4.366E+0 | 4.058E+1 | 2.566E+1 | 1.695E+2 | 1.831E+2 | 4.203E+1 | 3.179E+1 |
| N_corr | 4.646E+1 | 2.920E+1 | 1.668E+3 | 8.006E+2 | 4.275E+0 | 4.866E+0 | 4.281E+1 | 2.765E+1 | 1.676E+2 | 3.958E+2 | 4.756E+1 | 3.353E+1 |
| c [nM] | 2.811E+2 | 1.712E+2 | 5.025E+3 | 2.178E+3 | 2.013E+1 | 2.244E+1 | 2.524E+2 | 1.596E+2 | 7.864E+2 | 8.495E+2 | 2.161E+2 | 1.634E+2 |
| c_corr [nM] | 2.889E+2 | 1.816E+2 | 7.737E+3 | 3.714E+3 | 2.198E+1 | 2.502E+1 | 2.662E+2 | 1.720E+2 | 7.776E+2 | 1.836E+3 | 2.445E+2 | 1.724E+2 |
| ratioG | 5.737E-2 | 3.817E-2 | 6.257E-1 | 2.659E-1 | ||||||||
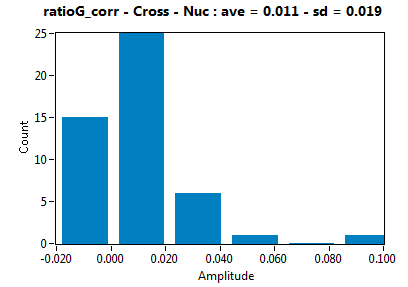
| ratioG_corr | 1.053E-2 | 1.928E-2 | 1.217E-1 | 1.197E-1 | ||||||||
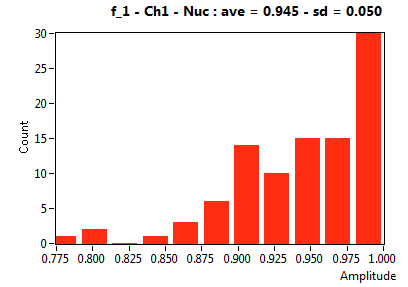
| f_1 | 9.453E-1 | 5.012E-2 | 4.841E-1 | 1.863E-1 | 9.864E-1 | 3.770E-2 | 9.317E-1 | 5.024E-2 | 9.731E-1 | 8.117E-2 | 9.599E-1 | 3.818E-2 |
| tauD_1 [ms] | 4.238E-1 | 2.314E-1 | 1.868E+0 | 8.939E-2 | 2.738E-1 | 1.647E-1 | 3.487E-1 | 1.920E-1 | 7.537E-1 | 6.216E-1 | 1.921E-1 | 6.166E-2 |
| D_1 [mum^2/s] | 3.115E+1 | 1.274E+1 | 6.912E+0 | 3.198E-1 | 5.987E+1 | 3.205E+1 | 3.554E+1 | 1.167E+1 | 3.735E+1 | 3.251E+1 | 6.915E+1 | 2.298E+1 |
| alpha_1 | 1.018E+0 | 7.306E-2 | 9.443E-1 | 3.681E-2 | 1.005E+0 | 2.024E-1 | 1.022E+0 | 1.043E-1 | 8.916E-1 | 2.200E-1 | 1.013E+0 | 2.002E-1 |
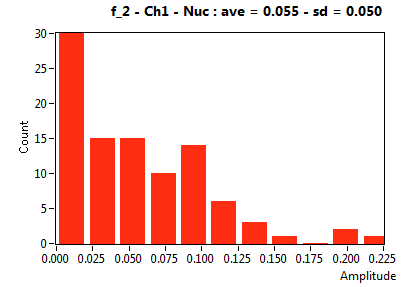
| f_2 | 5.467E-2 | 5.012E-2 | 5.159E-1 | 1.863E-1 | 1.359E-2 | 3.770E-2 | 6.831E-2 | 5.024E-2 | 2.690E-2 | 8.117E-2 | 4.012E-2 | 3.818E-2 |
| tauD_2 [ms] | 7.056E+0 | 3.207E+0 | 7.983E+1 | 9.256E+1 | 4.083E+1 | 2.404E+1 | 1.163E+1 | 1.321E+1 | 1.148E+1 | 1.486E+1 | 1.200E+1 | 7.499E+0 |
| D_2 [mum^2/s] | 1.726E+0 | 5.740E-1 | 3.936E-1 | 4.021E-1 | 3.908E-1 | 2.136E-1 | 1.491E+0 | 7.658E-1 | 2.132E+0 | 8.663E-1 | 1.369E+0 | 7.063E-1 |
| alpha_2 | 1.089E+0 | 1.091E-1 | 1.319E+0 | 3.289E-1 | 1.070E+0 | 5.368E-2 | 1.068E+0 | 1.396E-1 | 1.065E+0 | 1.927E-1 | 1.074E+0 | 9.195E-3 |
| tauD_m [ms] | 8.686E-1 | 7.820E-1 | 4.791E+1 | 7.057E+1 | 1.151E+0 | 3.989E+0 | 1.280E+0 | 1.724E+0 | 2.066E+0 | 4.243E+0 | 5.214E-1 | 3.679E-1 |
| D_m [mum^2/s] | 2.973E+1 | 1.280E+1 | 3.527E+0 | 1.229E+0 | 5.927E+1 | 3.229E+1 | 3.337E+1 | 1.134E+1 | 3.717E+1 | 3.269E+1 | 6.679E+1 | 2.377E+1 |
| R [1/s] | 2.080E+2 | 8.387E+1 | 5.252E+1 | 1.577E+2 | ||||||||
| CPM [kHz] | 1.316E+0 | 1.746E-1 | 2.919E-1 | 2.395E-1 | 1.231E+0 | 4.252E-1 | 1.463E-1 | 6.598E-2 | ||||
| CPM_corr [kHz] | 1.373E+0 | 1.827E-1 | 3.175E-1 | 2.619E-1 | 1.304E+0 | 4.179E-1 | 7.019E-1 | 3.914E-1 | ||||
| Int [kHz] | 6.999E+1 | 4.327E+1 | 1.684E+2 | 4.745E+1 | 5.894E+1 | 4.311E+1 | 3.215E+1 | 4.223E+1 | ||||
| Int_corr [kHz] | 7.387E+1 | 4.558E+1 | 2.760E+2 | 8.628E+1 | 6.413E+1 | 4.536E+1 | 4.910E+1 | 8.084E+1 | ||||
| # samples | 97 | 97 | 69 | 69 | 48 | 48 | 104 | 104 | 19 | 19 | 17 | 17 |
| # cells | 41 |
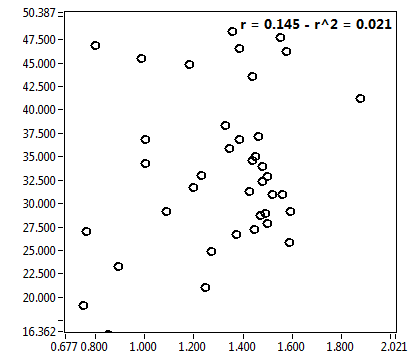
![c_corr [nM] - Ch1 - Nuc : ave = 288.928 - sd = 181.596](0592_c_corr_1n.png)
![Int_corr [kHz] - Ch1 - Nuc : ave = 73.874 - sd = 45.584](0592_Int_corr_1n.png)
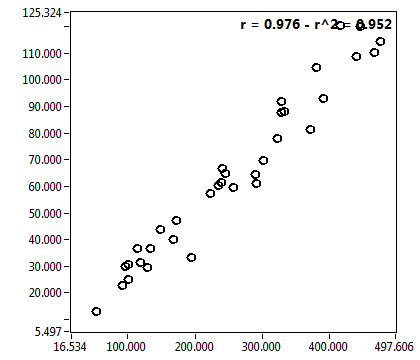
![D_1 [mum^2/s] - Ch1 - Nuc : ave = 31.152 - sd = 12.738](0592_D_1_1n.png)
![D_2 [mum^2/s] - Ch1 - Nuc : ave = 1.726 - sd = 0.574](0592_D_2_1n.png)
![CPM_corr [kHz] - Ch1 - Nuc : ave = 1.373 - sd = 0.183](0592_CPM_corr_1n.png)
![D_m [mum^2/s] - Ch1 - Nuc : ave = 29.734 - sd = 12.805](0592_D_m_1n.png)
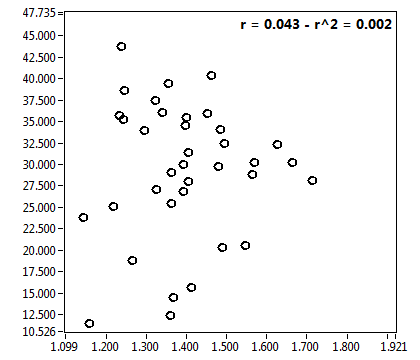
![c_corr [nM] - Ch1 - Nuc : ave = 288.928 - sd = 181.596](0592_c_corr_1n.png)
![c_corr [nM] - Ch1 - Cyto : ave = 266.216 - sd = 171.968](0592_c_corr_1c.png)
![Int_corr [kHz] - Ch1 - Cyto : ave = 64.127 - sd = 45.363](0592_Int_corr_1c.png)
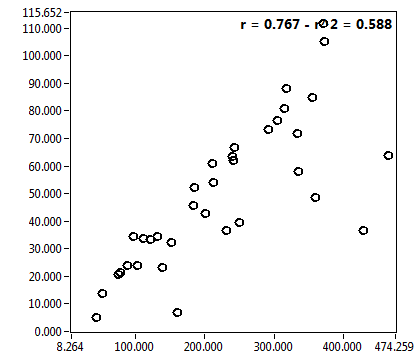
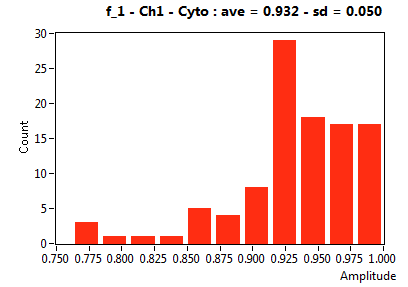
![D_1 [mum^2/s] - Ch1 - Cyto : ave = 35.538 - sd = 11.665](0592_D_1_1c.png)
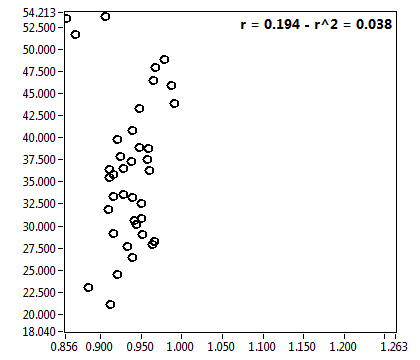
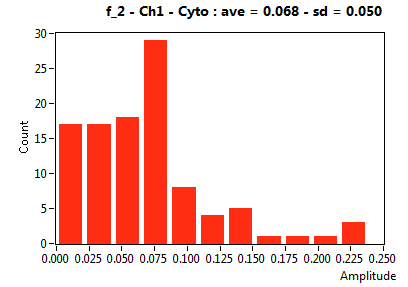
![D_2 [mum^2/s] - Ch1 - Cyto : ave = 1.491 - sd = 0.766](0592_D_2_1c.png)
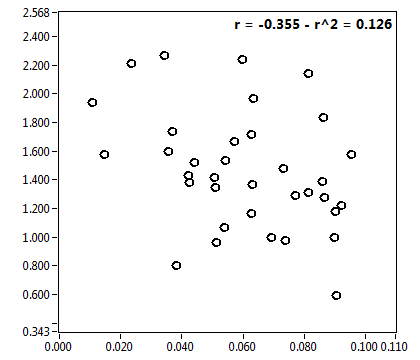
![CPM_corr [kHz] - Ch1 - Cyto : ave = 1.304 - sd = 0.418](0592_CPM_corr_1c.png)
![D_m [mum^2/s] - Ch1 - Cyto : ave = 33.372 - sd = 11.340](0592_D_m_1c.png)